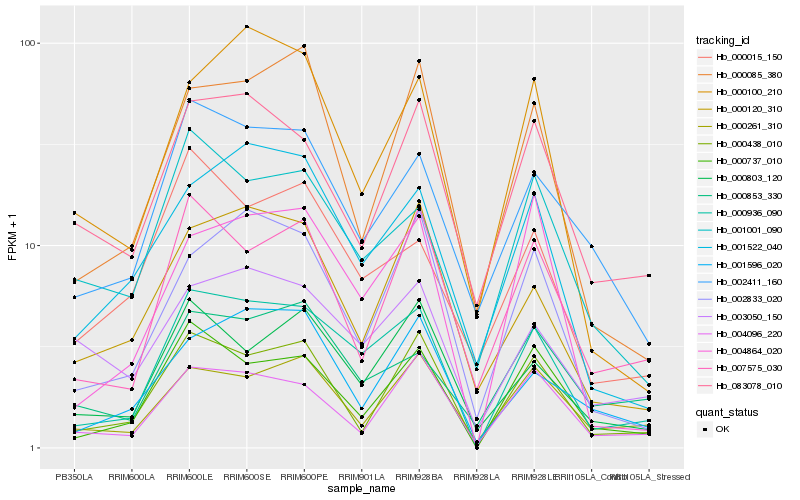
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000936_090 |
0.0 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RING1a isoform X2 [Jatropha curcas] |
| 2 |
Hb_000737_010 |
0.1353161135 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RING1a isoform X1 [Jatropha curcas] |
| 3 |
Hb_000438_010 |
0.1543621733 |
- |
- |
- |
| 4 |
Hb_004864_020 |
0.1553657761 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 5 |
Hb_001522_040 |
0.1594270006 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 6 |
Hb_002411_160 |
0.1609868948 |
- |
- |
PREDICTED: uncharacterized protein LOC105631262 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000803_120 |
0.161338161 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 22-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_002833_020 |
0.1635230048 |
desease resistance |
Gene Name: LRR_4 |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 9 |
Hb_001001_090 |
0.1685071065 |
- |
- |
PREDICTED: uncharacterized protein LOC105633658 [Jatropha curcas] |
| 10 |
Hb_007575_030 |
0.168876238 |
- |
- |
PREDICTED: protein RCC2 homolog [Jatropha curcas] |
| 11 |
Hb_000261_310 |
0.1692468656 |
- |
- |
PREDICTED: UV radiation resistance-associated gene protein isoform X3 [Jatropha curcas] |
| 12 |
Hb_004096_220 |
0.1698231193 |
- |
- |
PREDICTED: uncharacterized protein LOC105638706 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000120_310 |
0.1725106499 |
- |
- |
Gibberellin 3-beta-dioxygenase, putative [Ricinus communis] |
| 14 |
Hb_000853_330 |
0.1725163531 |
- |
- |
PREDICTED: importin subunit alpha-like [Jatropha curcas] |
| 15 |
Hb_001596_020 |
0.1727304307 |
- |
- |
hypothetical protein CICLE_v10014140mg [Citrus clementina] |
| 16 |
Hb_000015_150 |
0.1741045832 |
- |
- |
Ubiquitin-conjugating enzyme E2 7 family protein [Populus trichocarpa] |
| 17 |
Hb_000100_210 |
0.1746562479 |
- |
- |
PREDICTED: selenium-binding protein 1-like [Jatropha curcas] |
| 18 |
Hb_083078_010 |
0.1748190751 |
- |
- |
Splicing factor, arginine/serine-rich, putative [Ricinus communis] |
| 19 |
Hb_000085_380 |
0.175110974 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003050_150 |
0.1759207097 |
- |
- |
endoplasmic reticulum mannosyl-oligosaccharide 1,2-alpha-mannosidase, putative [Ricinus communis] |