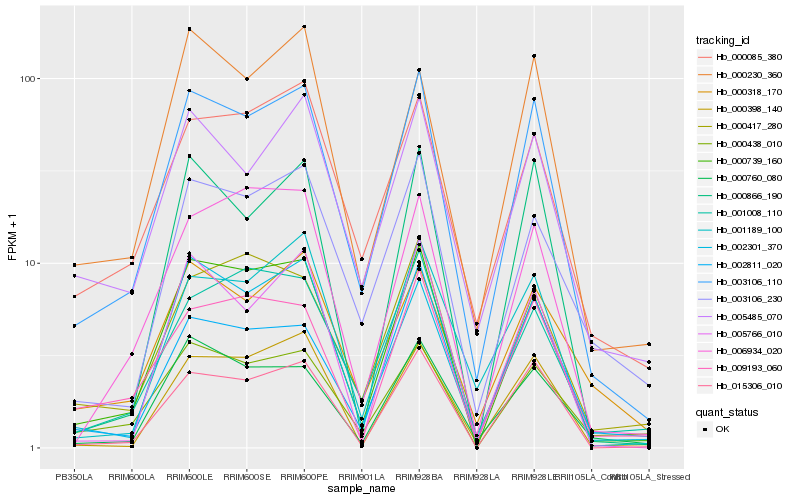
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003106_110 |
0.0 |
- |
- |
betaine aldehyde dehydrogenase 1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000739_160 |
0.0869610662 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_015306_010 |
0.0928667716 |
- |
- |
PREDICTED: uncharacterized protein LOC105636357 [Jatropha curcas] |
| 4 |
Hb_002301_370 |
0.1107798003 |
- |
- |
hypothetical protein JCGZ_21454 [Jatropha curcas] |
| 5 |
Hb_005485_070 |
0.1124064131 |
- |
- |
5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, putative [Ricinus communis] |
| 6 |
Hb_000085_380 |
0.1162763252 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000760_080 |
0.11735775 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 8 |
Hb_000398_140 |
0.1191380395 |
- |
- |
PREDICTED: methionine S-methyltransferase [Jatropha curcas] |
| 9 |
Hb_000438_010 |
0.1203146226 |
- |
- |
- |
| 10 |
Hb_009193_060 |
0.1203657996 |
- |
- |
PREDICTED: ureide permease 1-like [Jatropha curcas] |
| 11 |
Hb_003106_230 |
0.1206148208 |
transcription factor |
TF Family: LIM |
PREDICTED: LIM domain-containing protein WLIM1-like [Jatropha curcas] |
| 12 |
Hb_000230_360 |
0.1228846384 |
- |
- |
PREDICTED: BI1-like protein [Jatropha curcas] |
| 13 |
Hb_000318_170 |
0.1238225945 |
- |
- |
N-acetyl-gamma-glutamyl-phosphate reductase, putative [Ricinus communis] |
| 14 |
Hb_001189_100 |
0.1252837698 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 2-like [Jatropha curcas] |
| 15 |
Hb_001008_110 |
0.1271110473 |
- |
- |
- |
| 16 |
Hb_005766_010 |
0.1273479072 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 17 |
Hb_000866_190 |
0.127694987 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 18 |
Hb_000417_280 |
0.1279716542 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002811_020 |
0.1285337714 |
- |
- |
Cysteine synthase [Morus notabilis] |
| 20 |
Hb_006934_020 |
0.1301791065 |
- |
- |
hypothetical protein POPTR_0004s09051g [Populus trichocarpa] |