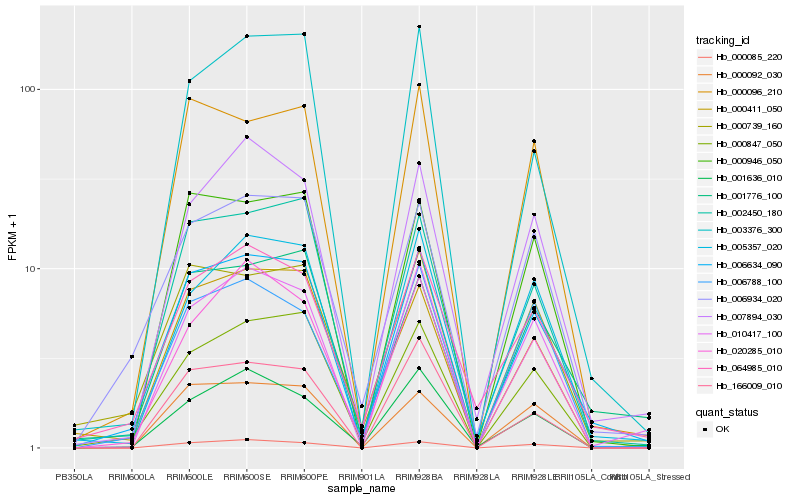
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006634_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646581 [Jatropha curcas] |
| 2 |
Hb_002450_180 |
0.0942055562 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000739_160 |
0.1042593066 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_000847_050 |
0.1042796033 |
- |
- |
PREDICTED: dCTP pyrophosphatase 1-like [Jatropha curcas] |
| 5 |
Hb_010417_100 |
0.1044563483 |
- |
- |
PREDICTED: respiratory burst oxidase homolog protein A [Jatropha curcas] |
| 6 |
Hb_000096_210 |
0.1065504274 |
- |
- |
PREDICTED: probable carboxylesterase 7 [Jatropha curcas] |
| 7 |
Hb_064985_010 |
0.1085292726 |
- |
- |
Leucine-rich repeat transmembrane protein kinase, putative [Theobroma cacao] |
| 8 |
Hb_001776_100 |
0.1086864041 |
- |
- |
PREDICTED: outer envelope pore protein 16, chloroplastic [Jatropha curcas] |
| 9 |
Hb_006934_020 |
0.1086868997 |
- |
- |
hypothetical protein POPTR_0004s09051g [Populus trichocarpa] |
| 10 |
Hb_005357_020 |
0.1106528881 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000411_050 |
0.112082752 |
- |
- |
hypothetical protein AMTR_s00066p00166230 [Amborella trichopoda] |
| 12 |
Hb_000946_050 |
0.1133266377 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 13 |
Hb_003376_300 |
0.11371658 |
- |
- |
Aquaporin PIP2.2, putative [Ricinus communis] |
| 14 |
Hb_007894_030 |
0.1169510145 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 15 |
Hb_000085_220 |
0.1208872369 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_006788_100 |
0.124199901 |
- |
- |
PREDICTED: uncharacterized protein LOC105649919 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001636_010 |
0.1264956089 |
- |
- |
hypothetical protein JCGZ_01264 [Jatropha curcas] |
| 18 |
Hb_000092_030 |
0.1272776997 |
- |
- |
PREDICTED: uncharacterized protein LOC105111213 [Populus euphratica] |
| 19 |
Hb_020285_010 |
0.1292453108 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3, partial [Pyrus x bretschneideri] |
| 20 |
Hb_166009_010 |
0.1294627534 |
- |
- |
carbonyl reductase, putative [Ricinus communis] |