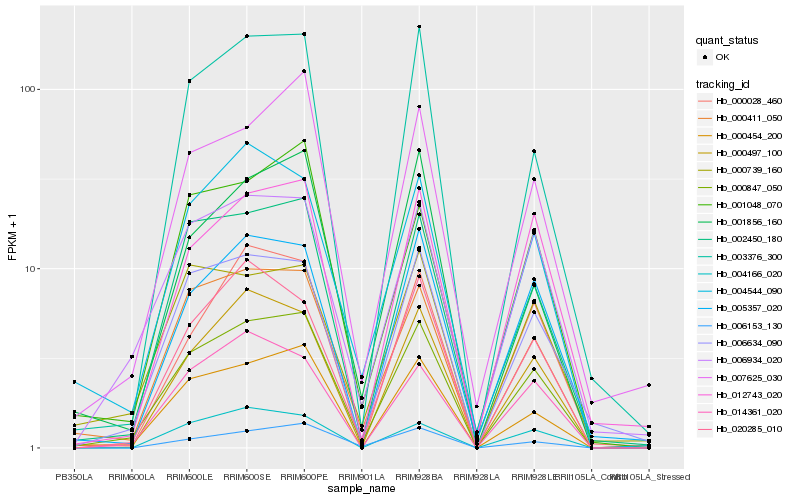
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000847_050 |
0.0 |
- |
- |
PREDICTED: dCTP pyrophosphatase 1-like [Jatropha curcas] |
| 2 |
Hb_000454_200 |
0.0849525452 |
- |
- |
PREDICTED: indole-3-acetic acid-induced protein ARG7 [Jatropha curcas] |
| 3 |
Hb_000411_050 |
0.0873531657 |
- |
- |
hypothetical protein AMTR_s00066p00166230 [Amborella trichopoda] |
| 4 |
Hb_002450_180 |
0.090314114 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_006934_020 |
0.1031194969 |
- |
- |
hypothetical protein POPTR_0004s09051g [Populus trichocarpa] |
| 6 |
Hb_006634_090 |
0.1042796033 |
- |
- |
PREDICTED: uncharacterized protein LOC105646581 [Jatropha curcas] |
| 7 |
Hb_000497_100 |
0.1087183467 |
- |
- |
respiratory burst oxidase D [Manihot esculenta] |
| 8 |
Hb_003376_300 |
0.1105666295 |
- |
- |
Aquaporin PIP2.2, putative [Ricinus communis] |
| 9 |
Hb_005357_020 |
0.1129469998 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_007625_030 |
0.1141505057 |
- |
- |
unknown [Medicago truncatula] |
| 11 |
Hb_004544_090 |
0.1174223199 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 12 |
Hb_012743_020 |
0.119335052 |
- |
- |
serine/threonine protein phosphatase, putative [Ricinus communis] |
| 13 |
Hb_006153_130 |
0.1197947876 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 14 |
Hb_001048_070 |
0.123334397 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0007s14590g [Populus trichocarpa] |
| 15 |
Hb_020285_010 |
0.1249730891 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3, partial [Pyrus x bretschneideri] |
| 16 |
Hb_014361_020 |
0.126211388 |
- |
- |
PREDICTED: probable protein phosphatase 2C 78 [Jatropha curcas] |
| 17 |
Hb_004166_020 |
0.1285533017 |
- |
- |
- |
| 18 |
Hb_000739_160 |
0.128859268 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_001856_160 |
0.1296599988 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000028_460 |
0.1307100457 |
- |
- |
hypothetical protein POPTR_0015s05340g [Populus trichocarpa] |