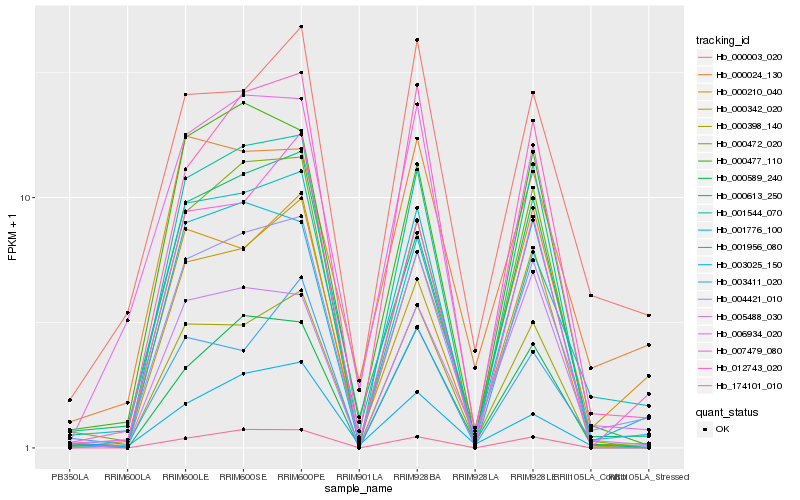
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004421_010 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 2 |
Hb_000398_140 |
0.0821849028 |
- |
- |
PREDICTED: methionine S-methyltransferase [Jatropha curcas] |
| 3 |
Hb_001776_100 |
0.0845224538 |
- |
- |
PREDICTED: outer envelope pore protein 16, chloroplastic [Jatropha curcas] |
| 4 |
Hb_001956_080 |
0.0920592859 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 14 [Jatropha curcas] |
| 5 |
Hb_000613_250 |
0.092774387 |
- |
- |
PREDICTED: nudix hydrolase 2-like [Jatropha curcas] |
| 6 |
Hb_012743_020 |
0.1049731807 |
- |
- |
serine/threonine protein phosphatase, putative [Ricinus communis] |
| 7 |
Hb_000472_020 |
0.1089835969 |
- |
- |
PREDICTED: uncharacterized protein LOC105641103 [Jatropha curcas] |
| 8 |
Hb_000477_110 |
0.110152218 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein SLR1-like [Jatropha curcas] |
| 9 |
Hb_006934_020 |
0.1144705647 |
- |
- |
hypothetical protein POPTR_0004s09051g [Populus trichocarpa] |
| 10 |
Hb_007479_080 |
0.115166097 |
- |
- |
PREDICTED: uncharacterized protein LOC105649774 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001544_070 |
0.1159575222 |
- |
- |
PREDICTED: multicopper oxidase LPR1 [Jatropha curcas] |
| 12 |
Hb_000210_040 |
0.1175665043 |
- |
- |
PREDICTED: putative HVA22-like protein g [Jatropha curcas] |
| 13 |
Hb_000342_020 |
0.1176606308 |
- |
- |
bel1 homeotic protein, putative [Ricinus communis] |
| 14 |
Hb_000589_240 |
0.121021666 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 15 |
Hb_000003_020 |
0.1250394886 |
- |
- |
PREDICTED: uncharacterized protein LOC105631161 [Jatropha curcas] |
| 16 |
Hb_005488_030 |
0.1258352028 |
- |
- |
F3F9.11 [Arabidopsis lyrata subsp. lyrata] |
| 17 |
Hb_000024_130 |
0.1278000296 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003411_020 |
0.1281432902 |
- |
- |
alpha-glucosidase, putative [Ricinus communis] |
| 19 |
Hb_003025_150 |
0.1294324467 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_174101_010 |
0.1298351806 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |