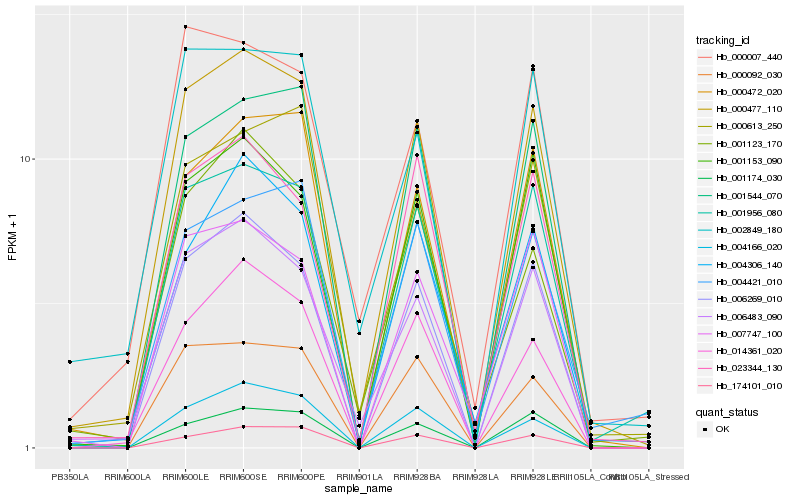
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000477_110 |
0.0 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein SLR1-like [Jatropha curcas] |
| 2 |
Hb_006483_090 |
0.0758489064 |
- |
- |
PREDICTED: uncharacterized protein LOC105639979 [Jatropha curcas] |
| 3 |
Hb_000472_020 |
0.0843398309 |
- |
- |
PREDICTED: uncharacterized protein LOC105641103 [Jatropha curcas] |
| 4 |
Hb_006269_010 |
0.0881195711 |
- |
- |
hypothetical protein JCGZ_21552 [Jatropha curcas] |
| 5 |
Hb_001956_080 |
0.0899528247 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 14 [Jatropha curcas] |
| 6 |
Hb_001153_090 |
0.0928130845 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 7 |
Hb_000613_250 |
0.0931844973 |
- |
- |
PREDICTED: nudix hydrolase 2-like [Jatropha curcas] |
| 8 |
Hb_004306_140 |
0.1024944814 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 9 |
Hb_001123_170 |
0.103213504 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_014361_020 |
0.1033458785 |
- |
- |
PREDICTED: probable protein phosphatase 2C 78 [Jatropha curcas] |
| 11 |
Hb_001544_070 |
0.1033686866 |
- |
- |
PREDICTED: multicopper oxidase LPR1 [Jatropha curcas] |
| 12 |
Hb_023344_130 |
0.1053102044 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 13 |
Hb_000092_030 |
0.1054602855 |
- |
- |
PREDICTED: uncharacterized protein LOC105111213 [Populus euphratica] |
| 14 |
Hb_004166_020 |
0.1091199418 |
- |
- |
- |
| 15 |
Hb_004421_010 |
0.110152218 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 16 |
Hb_002849_180 |
0.1108328871 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 17 |
Hb_001174_030 |
0.1117733102 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_007747_100 |
0.1140397249 |
- |
- |
PREDICTED: probable F-box protein At1g60180 [Jatropha curcas] |
| 19 |
Hb_174101_010 |
0.1142322022 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 20 |
Hb_000007_440 |
0.1150413066 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |