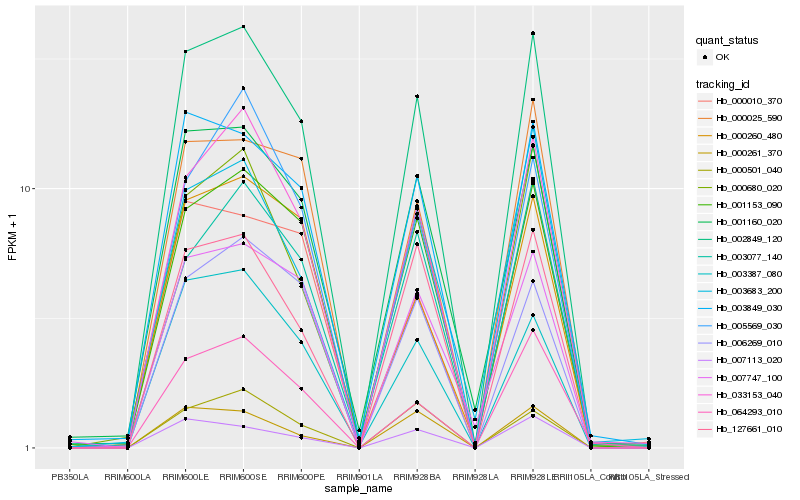
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002849_120 |
0.0 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 2 |
Hb_033153_040 |
0.0692826327 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 3 |
Hb_001153_090 |
0.0752207693 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 4 |
Hb_003849_030 |
0.0762409832 |
- |
- |
hypothetical protein AALP_AA5G029900 [Arabis alpina] |
| 5 |
Hb_000680_020 |
0.0845025332 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 6 |
Hb_001160_020 |
0.0857932634 |
transcription factor |
TF Family: NF-YA |
Nuclear transcription factor Y subunit A-3, putative [Ricinus communis] |
| 7 |
Hb_127661_010 |
0.0965427638 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_003387_080 |
0.0967846898 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_007747_100 |
0.1019119795 |
- |
- |
PREDICTED: probable F-box protein At1g60180 [Jatropha curcas] |
| 10 |
Hb_000025_590 |
0.1038655542 |
- |
- |
PREDICTED: cytochrome P450 94A1-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_007113_020 |
0.1041269784 |
- |
- |
- |
| 12 |
Hb_006269_010 |
0.106019606 |
- |
- |
hypothetical protein JCGZ_21552 [Jatropha curcas] |
| 13 |
Hb_003077_140 |
0.1062080264 |
- |
- |
hypothetical protein JCGZ_16294 [Jatropha curcas] |
| 14 |
Hb_064293_010 |
0.1064970892 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 15 |
Hb_003683_200 |
0.1103896862 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 16 |
Hb_000501_040 |
0.1177970881 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_000261_370 |
0.1185966388 |
- |
- |
- |
| 18 |
Hb_005569_030 |
0.1188772 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g56140 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000260_480 |
0.1191585487 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SCARECROW [Jatropha curcas] |
| 20 |
Hb_000010_370 |
0.1215925824 |
- |
- |
PREDICTED: uncharacterized protein LOC105640263 [Jatropha curcas] |