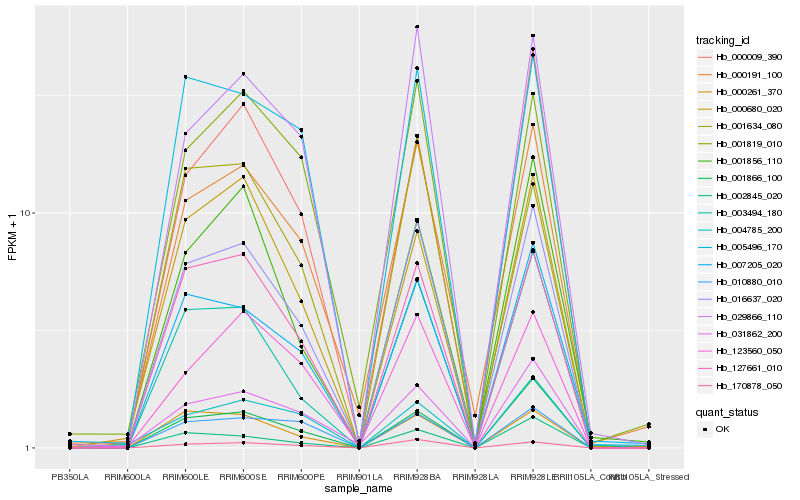
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_016637_020 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 2 |
Hb_127661_010 |
0.0838329727 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_000191_100 |
0.0848432022 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 4 |
Hb_003494_180 |
0.087612122 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 5 |
Hb_007205_020 |
0.0903000778 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 6 |
Hb_170878_050 |
0.0910198949 |
- |
- |
retrotransposon protein, putative, unclassified [Oryza sativa Japonica Group] |
| 7 |
Hb_029866_110 |
0.094025993 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 10-like [Jatropha curcas] |
| 8 |
Hb_031862_200 |
0.0961984291 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At3g14460 [Vitis vinifera] |
| 9 |
Hb_005496_170 |
0.1012198173 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 10 |
Hb_000261_370 |
0.10291588 |
- |
- |
- |
| 11 |
Hb_001866_100 |
0.1069309595 |
- |
- |
PREDICTED: uncharacterized protein LOC104212668 [Nicotiana sylvestris] |
| 12 |
Hb_002845_020 |
0.1094517627 |
- |
- |
hypothetical protein POPTR_0012s14920g [Populus trichocarpa] |
| 13 |
Hb_001819_010 |
0.1127045879 |
- |
- |
PREDICTED: potassium transporter 8 [Jatropha curcas] |
| 14 |
Hb_123560_050 |
0.1174638131 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g24010 [Jatropha curcas] |
| 15 |
Hb_010880_010 |
0.1223398883 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000680_020 |
0.123644423 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 17 |
Hb_000009_390 |
0.126308728 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_004785_200 |
0.1280610353 |
- |
- |
PREDICTED: protein MKS1 [Jatropha curcas] |
| 19 |
Hb_001634_080 |
0.1289242597 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 20 |
Hb_001856_110 |
0.1293142247 |
- |
- |
PREDICTED: cellulose synthase-like protein G2 [Populus euphratica] |