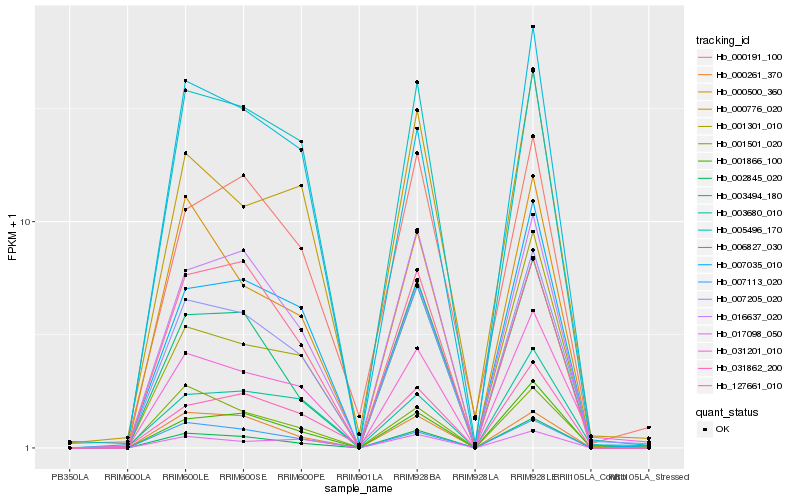
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007205_020 |
0.0 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 2 |
Hb_031201_010 |
0.0694890973 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_002845_020 |
0.0843695628 |
- |
- |
hypothetical protein POPTR_0012s14920g [Populus trichocarpa] |
| 4 |
Hb_016637_020 |
0.0903000778 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 5 |
Hb_003494_180 |
0.0957708111 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 6 |
Hb_001866_100 |
0.0972192115 |
- |
- |
PREDICTED: uncharacterized protein LOC104212668 [Nicotiana sylvestris] |
| 7 |
Hb_005496_170 |
0.1021103548 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 8 |
Hb_007035_010 |
0.1094757681 |
- |
- |
PREDICTED: synaptotagmin-4 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000191_100 |
0.1108775642 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 10 |
Hb_031862_200 |
0.1148605005 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At3g14460 [Vitis vinifera] |
| 11 |
Hb_000776_020 |
0.1156947746 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_25594 [Jatropha curcas] |
| 12 |
Hb_006827_030 |
0.1158457565 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 13 |
Hb_001301_010 |
0.1178342212 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 14 |
Hb_017098_050 |
0.1193235822 |
- |
- |
hypothetical protein POPTR_0001s03540g [Populus trichocarpa] |
| 15 |
Hb_007113_020 |
0.1201284576 |
- |
- |
- |
| 16 |
Hb_001501_020 |
0.1213040188 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 17 |
Hb_000500_360 |
0.1229539894 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 18 |
Hb_127661_010 |
0.1236773591 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_003680_010 |
0.1242120146 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 20 |
Hb_000261_370 |
0.1244373985 |
- |
- |
- |