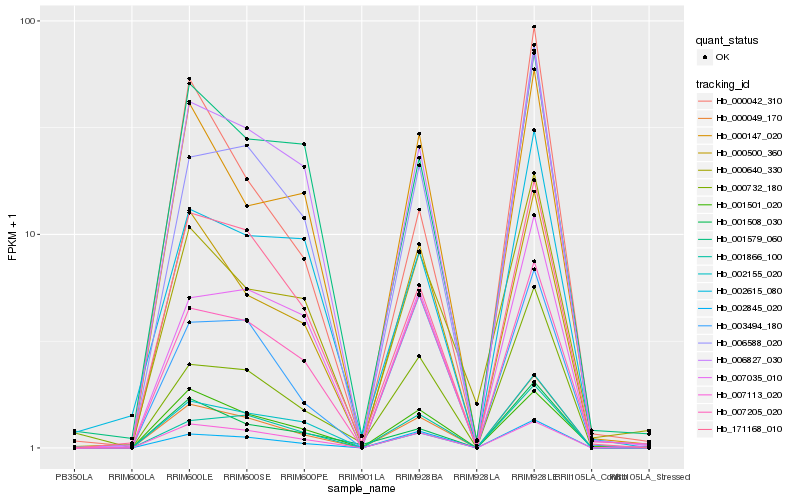
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000049_170 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002845_020 |
0.0886984943 |
- |
- |
hypothetical protein POPTR_0012s14920g [Populus trichocarpa] |
| 3 |
Hb_006827_030 |
0.090198023 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 4 |
Hb_006588_020 |
0.0977801977 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_001866_100 |
0.1028625778 |
- |
- |
PREDICTED: uncharacterized protein LOC104212668 [Nicotiana sylvestris] |
| 6 |
Hb_000500_360 |
0.1065818538 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_171168_010 |
0.1113279868 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 8 |
Hb_000147_020 |
0.122364065 |
- |
- |
PREDICTED: uncharacterized protein LOC105638303 [Jatropha curcas] |
| 9 |
Hb_007035_010 |
0.1225160213 |
- |
- |
PREDICTED: synaptotagmin-4 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001508_030 |
0.1259940495 |
- |
- |
hypothetical protein VITISV_029808 [Vitis vinifera] |
| 11 |
Hb_007113_020 |
0.1262178723 |
- |
- |
- |
| 12 |
Hb_007205_020 |
0.1285880717 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 13 |
Hb_000640_330 |
0.1310651722 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000732_180 |
0.1360716621 |
- |
- |
Uncharacterized protein TCM_030007 [Theobroma cacao] |
| 15 |
Hb_001579_060 |
0.1373209224 |
- |
- |
carbonic anhydrase, putative [Ricinus communis] |
| 16 |
Hb_003494_180 |
0.1395381584 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 17 |
Hb_002615_080 |
0.1420034222 |
- |
- |
PREDICTED: non-specific phospholipase C2 [Jatropha curcas] |
| 18 |
Hb_002155_020 |
0.1424995694 |
- |
- |
PREDICTED: probable WRKY transcription factor 65 [Jatropha curcas] |
| 19 |
Hb_000042_310 |
0.1493031869 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 20 |
Hb_001501_020 |
0.1495388774 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |