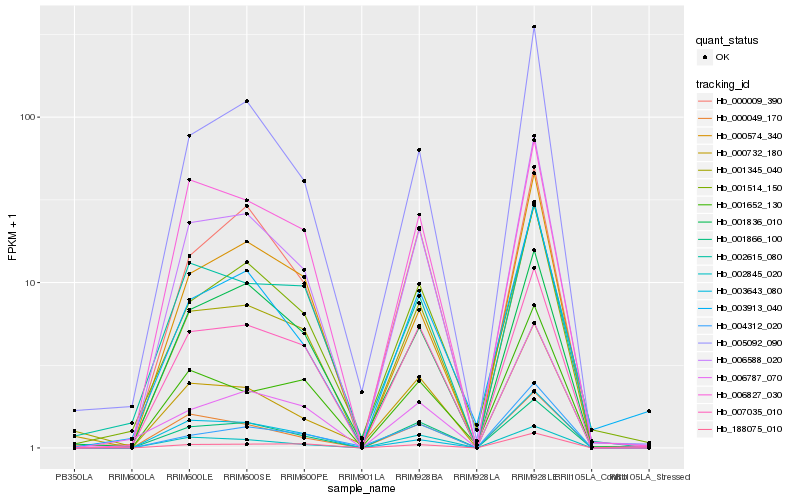
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006588_020 |
0.0 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_005092_090 |
0.0789873957 |
- |
- |
cold regulated protein [Manihot esculenta] |
| 3 |
Hb_001866_100 |
0.0840411072 |
- |
- |
PREDICTED: uncharacterized protein LOC104212668 [Nicotiana sylvestris] |
| 4 |
Hb_001345_040 |
0.0885529798 |
- |
- |
hypothetical protein VITISV_015004 [Vitis vinifera] |
| 5 |
Hb_007035_010 |
0.091973671 |
- |
- |
PREDICTED: synaptotagmin-4 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000049_170 |
0.0977801977 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000009_390 |
0.1061022095 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001514_150 |
0.1077865524 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 9 |
Hb_188075_010 |
0.110276848 |
- |
- |
PREDICTED: uncharacterized protein LOC105775124 [Gossypium raimondii] |
| 10 |
Hb_003913_040 |
0.1138309026 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 11 |
Hb_000574_340 |
0.1145250382 |
- |
- |
hypothetical protein RCOM_0901420 [Ricinus communis] |
| 12 |
Hb_006827_030 |
0.1158656959 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 13 |
Hb_001836_010 |
0.1165100318 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g03230 [Jatropha curcas] |
| 14 |
Hb_002615_080 |
0.1240630639 |
- |
- |
PREDICTED: non-specific phospholipase C2 [Jatropha curcas] |
| 15 |
Hb_001652_130 |
0.1252157221 |
- |
- |
hypothetical protein B456_002G193900 [Gossypium raimondii] |
| 16 |
Hb_000732_180 |
0.1269270849 |
- |
- |
Uncharacterized protein TCM_030007 [Theobroma cacao] |
| 17 |
Hb_002845_020 |
0.127425673 |
- |
- |
hypothetical protein POPTR_0012s14920g [Populus trichocarpa] |
| 18 |
Hb_004312_020 |
0.1292333414 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003643_080 |
0.1317374734 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 20 |
Hb_006787_070 |
0.131990512 |
- |
- |
PREDICTED: uncharacterized protein LOC104121706, partial [Nicotiana tomentosiformis] |