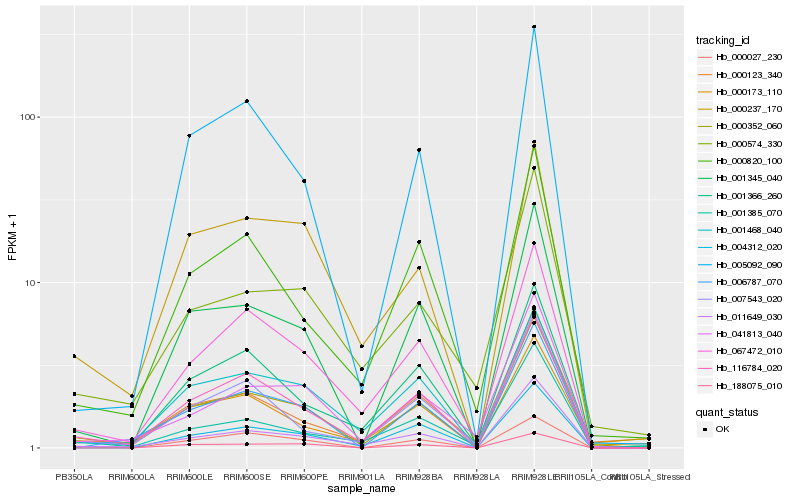
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000352_060 |
0.0 |
- |
- |
RNA-directed DNA polymerase [Arachis hypogaea] |
| 2 |
Hb_116784_020 |
0.0900730965 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 3 |
Hb_011649_030 |
0.0996168817 |
- |
- |
hypothetical protein [Vitis hybrid cultivar] |
| 4 |
Hb_001345_040 |
0.104097178 |
- |
- |
hypothetical protein VITISV_015004 [Vitis vinifera] |
| 5 |
Hb_000173_110 |
0.1180024989 |
- |
- |
putative RNA-directed DNA polymerase (Reverse transcriptase) [Malus domestica] |
| 6 |
Hb_067472_010 |
0.1235495404 |
- |
- |
Uncharacterized protein TCM_024268 [Theobroma cacao] |
| 7 |
Hb_001468_040 |
0.1258925387 |
- |
- |
Uncharacterized protein TCM_024268 [Theobroma cacao] |
| 8 |
Hb_006787_070 |
0.1272118449 |
- |
- |
PREDICTED: uncharacterized protein LOC104121706, partial [Nicotiana tomentosiformis] |
| 9 |
Hb_000820_100 |
0.1288242656 |
- |
- |
RNA-directed DNA polymerase [Arachis hypogaea] |
| 10 |
Hb_001366_260 |
0.1301424945 |
- |
- |
T4.15 [Malus x robusta] |
| 11 |
Hb_000027_230 |
0.1301644221 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 12 |
Hb_041813_040 |
0.1303309247 |
- |
- |
NADH2 dehydrogenase (ubiquinone) (EC 1.6.5.3) chain 5, truncated - sugar beet mitochondrion (mitochondrion) [Beta vulgaris subsp. vulgaris] |
| 13 |
Hb_005092_090 |
0.1327292277 |
- |
- |
cold regulated protein [Manihot esculenta] |
| 14 |
Hb_007543_020 |
0.1342256984 |
- |
- |
PREDICTED: uncharacterized protein LOC104229533, partial [Nicotiana sylvestris] |
| 15 |
Hb_000123_340 |
0.1350169837 |
- |
- |
PREDICTED: uncharacterized protein LOC105789586 [Gossypium raimondii] |
| 16 |
Hb_004312_020 |
0.1368906161 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000574_330 |
0.1389729157 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 18 |
Hb_001385_070 |
0.1398586778 |
- |
- |
hypothetical protein VITISV_019194 [Vitis vinifera] |
| 19 |
Hb_000237_170 |
0.1437901908 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 6 [Jatropha curcas] |
| 20 |
Hb_188075_010 |
0.1464834609 |
- |
- |
PREDICTED: uncharacterized protein LOC105775124 [Gossypium raimondii] |