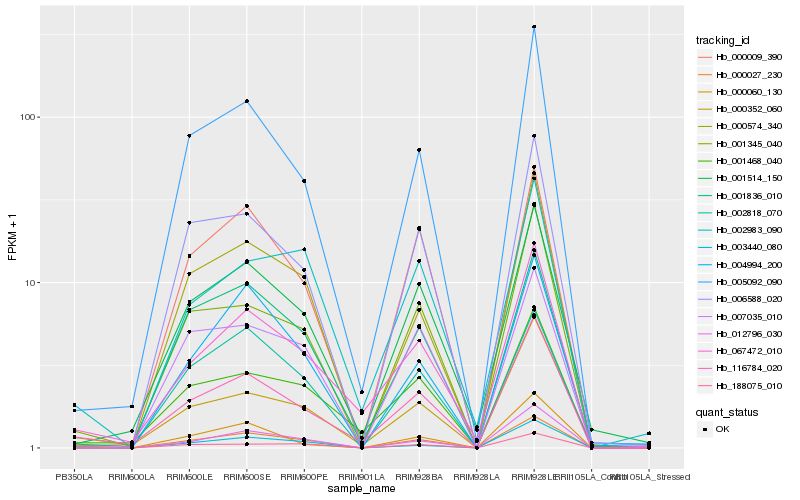
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000027_230 |
0.0 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 2 |
Hb_116784_020 |
0.1001330484 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 3 |
Hb_000574_340 |
0.1211479234 |
- |
- |
hypothetical protein RCOM_0901420 [Ricinus communis] |
| 4 |
Hb_005092_090 |
0.1227193425 |
- |
- |
cold regulated protein [Manihot esculenta] |
| 5 |
Hb_001345_040 |
0.1236165982 |
- |
- |
hypothetical protein VITISV_015004 [Vitis vinifera] |
| 6 |
Hb_067472_010 |
0.1293700613 |
- |
- |
Uncharacterized protein TCM_024268 [Theobroma cacao] |
| 7 |
Hb_000352_060 |
0.1301644221 |
- |
- |
RNA-directed DNA polymerase [Arachis hypogaea] |
| 8 |
Hb_001836_010 |
0.1337341191 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g03230 [Jatropha curcas] |
| 9 |
Hb_006588_020 |
0.1339651635 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_004994_200 |
0.135349129 |
- |
- |
PREDICTED: uncharacterized protein LOC105174340 [Sesamum indicum] |
| 11 |
Hb_012796_030 |
0.135662526 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 12 |
Hb_001514_150 |
0.1368930212 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 13 |
Hb_188075_010 |
0.1403434935 |
- |
- |
PREDICTED: uncharacterized protein LOC105775124 [Gossypium raimondii] |
| 14 |
Hb_000009_390 |
0.1408736109 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001468_040 |
0.1412612435 |
- |
- |
Uncharacterized protein TCM_024268 [Theobroma cacao] |
| 16 |
Hb_002983_090 |
0.1452815233 |
- |
- |
Altered inheritance of mitochondria protein 32 [Glycine soja] |
| 17 |
Hb_002818_070 |
0.1459759864 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 18 |
Hb_003440_080 |
0.1488918105 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 19 |
Hb_007035_010 |
0.1494111325 |
- |
- |
PREDICTED: synaptotagmin-4 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000060_130 |
0.1496002567 |
- |
- |
- |