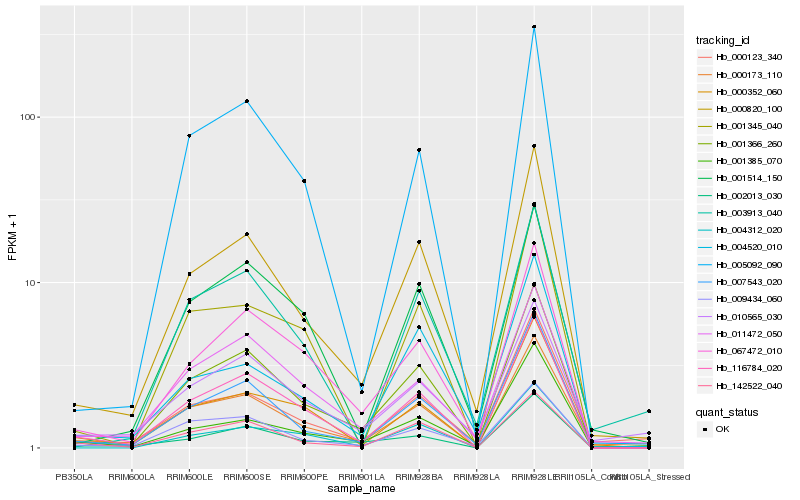
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000820_100 |
0.0 |
- |
- |
RNA-directed DNA polymerase [Arachis hypogaea] |
| 2 |
Hb_001366_260 |
0.0523478206 |
- |
- |
T4.15 [Malus x robusta] |
| 3 |
Hb_007543_020 |
0.0627795157 |
- |
- |
PREDICTED: uncharacterized protein LOC104229533, partial [Nicotiana sylvestris] |
| 4 |
Hb_004520_010 |
0.098549671 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 5 |
Hb_000173_110 |
0.0990336043 |
- |
- |
putative RNA-directed DNA polymerase (Reverse transcriptase) [Malus domestica] |
| 6 |
Hb_116784_020 |
0.1012855849 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 7 |
Hb_000123_340 |
0.1039889114 |
- |
- |
PREDICTED: uncharacterized protein LOC105789586 [Gossypium raimondii] |
| 8 |
Hb_067472_010 |
0.1095112292 |
- |
- |
Uncharacterized protein TCM_024268 [Theobroma cacao] |
| 9 |
Hb_009434_060 |
0.1116939603 |
- |
- |
T4.15 [Malus x robusta] |
| 10 |
Hb_142522_040 |
0.1209317688 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 11 |
Hb_001385_070 |
0.1236829064 |
- |
- |
hypothetical protein VITISV_019194 [Vitis vinifera] |
| 12 |
Hb_004312_020 |
0.1267376479 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005092_090 |
0.1271366302 |
- |
- |
cold regulated protein [Manihot esculenta] |
| 14 |
Hb_011472_050 |
0.1280732292 |
- |
- |
- |
| 15 |
Hb_003913_040 |
0.128103893 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 16 |
Hb_000352_060 |
0.1288242656 |
- |
- |
RNA-directed DNA polymerase [Arachis hypogaea] |
| 17 |
Hb_001514_150 |
0.1302752795 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 18 |
Hb_010565_030 |
0.1310814711 |
- |
- |
hypothetical protein VITISV_006810 [Vitis vinifera] |
| 19 |
Hb_002013_030 |
0.1345893584 |
- |
- |
PREDICTED: uncharacterized protein LOC104799692 [Tarenaya hassleriana] |
| 20 |
Hb_001345_040 |
0.1350429176 |
- |
- |
hypothetical protein VITISV_015004 [Vitis vinifera] |