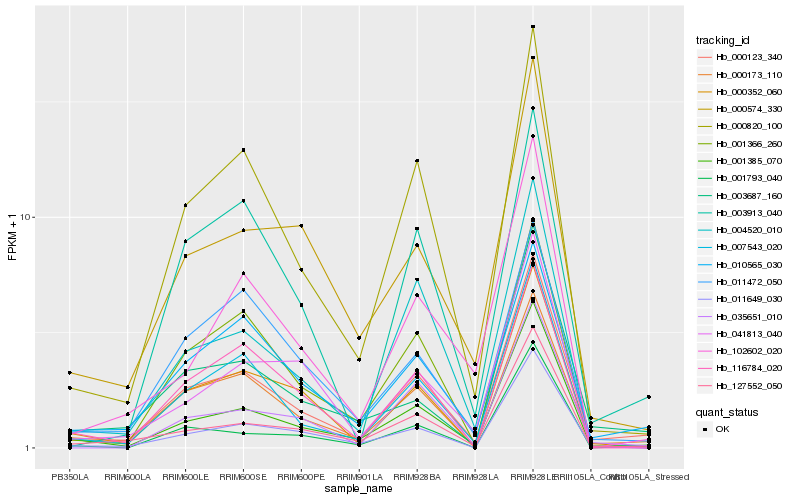
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000123_340 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105789586 [Gossypium raimondii] |
| 2 |
Hb_001385_070 |
0.094925595 |
- |
- |
hypothetical protein VITISV_019194 [Vitis vinifera] |
| 3 |
Hb_000173_110 |
0.0964639276 |
- |
- |
putative RNA-directed DNA polymerase (Reverse transcriptase) [Malus domestica] |
| 4 |
Hb_000820_100 |
0.1039889114 |
- |
- |
RNA-directed DNA polymerase [Arachis hypogaea] |
| 5 |
Hb_127552_050 |
0.1115979516 |
- |
- |
hypothetical protein VITISV_030752 [Vitis vinifera] |
| 6 |
Hb_007543_020 |
0.119123967 |
- |
- |
PREDICTED: uncharacterized protein LOC104229533, partial [Nicotiana sylvestris] |
| 7 |
Hb_010565_030 |
0.1253222308 |
- |
- |
hypothetical protein VITISV_006810 [Vitis vinifera] |
| 8 |
Hb_003687_160 |
0.1258535564 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 9 |
Hb_011649_030 |
0.1275934102 |
- |
- |
hypothetical protein [Vitis hybrid cultivar] |
| 10 |
Hb_004520_010 |
0.1292281181 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 11 |
Hb_001366_260 |
0.1299816221 |
- |
- |
T4.15 [Malus x robusta] |
| 12 |
Hb_000352_060 |
0.1350169837 |
- |
- |
RNA-directed DNA polymerase [Arachis hypogaea] |
| 13 |
Hb_000574_330 |
0.1382573152 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 14 |
Hb_035651_010 |
0.1386006895 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 15 |
Hb_003913_040 |
0.1477305948 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 16 |
Hb_001793_040 |
0.1483225012 |
- |
- |
hypothetical protein VITISV_006810 [Vitis vinifera] |
| 17 |
Hb_011472_050 |
0.153754999 |
- |
- |
- |
| 18 |
Hb_102602_020 |
0.1557994984 |
- |
- |
hypothetical protein B456_001G166300 [Gossypium raimondii] |
| 19 |
Hb_116784_020 |
0.1565979335 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 20 |
Hb_041813_040 |
0.1590995217 |
- |
- |
NADH2 dehydrogenase (ubiquinone) (EC 1.6.5.3) chain 5, truncated - sugar beet mitochondrion (mitochondrion) [Beta vulgaris subsp. vulgaris] |