| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
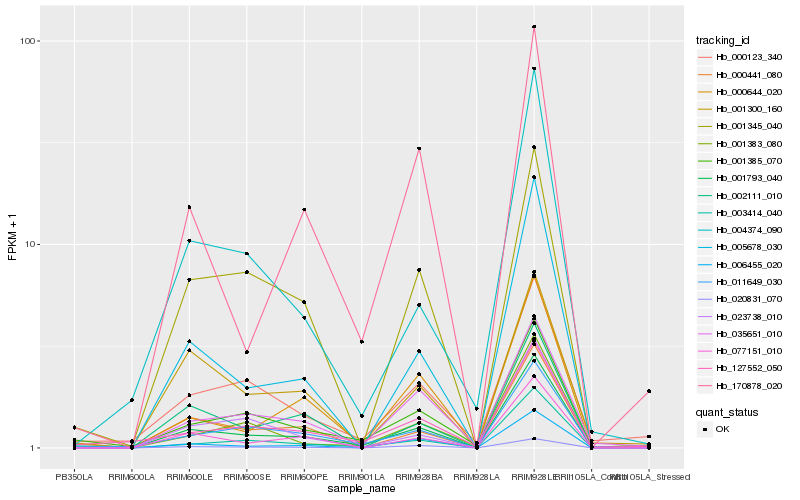
Hb_001793_040 |
0.0 |
- |
- |
hypothetical protein VITISV_006810 [Vitis vinifera] |
| 2 |
Hb_001385_070 |
0.1141512775 |
- |
- |
hypothetical protein VITISV_019194 [Vitis vinifera] |
| 3 |
Hb_003414_040 |
0.1305164871 |
- |
- |
hypothetical protein VITISV_016691 [Vitis vinifera] |
| 4 |
Hb_005678_030 |
0.1333857818 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 5 |
Hb_011649_030 |
0.1364040557 |
- |
- |
hypothetical protein [Vitis hybrid cultivar] |
| 6 |
Hb_127552_050 |
0.1417493863 |
- |
- |
hypothetical protein VITISV_030752 [Vitis vinifera] |
| 7 |
Hb_020831_070 |
0.143742127 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 8 |
Hb_077151_010 |
0.1457175425 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 9 |
Hb_000441_080 |
0.1480904004 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 10 |
Hb_000123_340 |
0.1483225012 |
- |
- |
PREDICTED: uncharacterized protein LOC105789586 [Gossypium raimondii] |
| 11 |
Hb_000644_020 |
0.1522584061 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 12 |
Hb_170878_020 |
0.1525040442 |
- |
- |
- |
| 13 |
Hb_006455_020 |
0.1558580978 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_035651_010 |
0.1559813259 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 15 |
Hb_023738_010 |
0.1561852744 |
- |
- |
Concanavalin A-like lectin protein kinase family protein [Theobroma cacao] |
| 16 |
Hb_002111_010 |
0.1581258005 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 17 |
Hb_004374_090 |
0.1602831666 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein BEL1 homolog [Populus euphratica] |
| 18 |
Hb_001300_160 |
0.1611252743 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 2-like [Jatropha curcas] |
| 19 |
Hb_001383_080 |
0.1624631718 |
- |
- |
OSJNBa0019D11.8 [Oryza sativa Japonica Group] |
| 20 |
Hb_001345_040 |
0.1653546571 |
- |
- |
hypothetical protein VITISV_015004 [Vitis vinifera] |