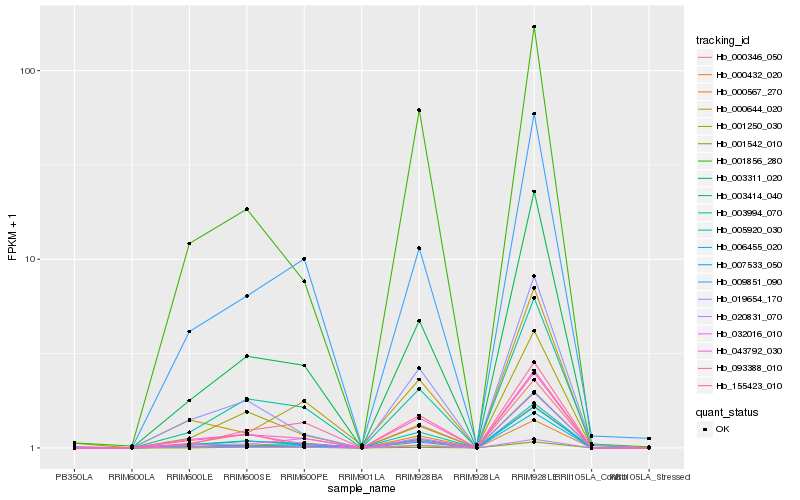
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003311_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003994_070 |
0.0842866886 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 3 |
Hb_000567_270 |
0.0859016854 |
- |
- |
hypothetical protein L484_017162 [Morus notabilis] |
| 4 |
Hb_032016_010 |
0.0911076852 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At3g28040-like isoform X1 [Citrus sinensis] |
| 5 |
Hb_009851_090 |
0.0920416506 |
- |
- |
Protein ABIL2, putative [Ricinus communis] |
| 6 |
Hb_007533_050 |
0.1011240916 |
- |
- |
- |
| 7 |
Hb_000432_020 |
0.1123142336 |
desease resistance |
Gene Name: Tropomyosin_1 |
PREDICTED: putative disease resistance protein RGA1 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001250_030 |
0.1153520892 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_005920_030 |
0.1167303598 |
- |
- |
putative polyprotein [Oryza sativa Japonica Group] |
| 10 |
Hb_019654_170 |
0.1193094698 |
transcription factor |
TF Family: ARID |
PREDICTED: high mobility group B protein 9 [Jatropha curcas] |
| 11 |
Hb_003414_040 |
0.1209696158 |
- |
- |
hypothetical protein VITISV_016691 [Vitis vinifera] |
| 12 |
Hb_155423_010 |
0.1222486839 |
- |
- |
- |
| 13 |
Hb_043792_030 |
0.122291701 |
- |
- |
PREDICTED: uncharacterized protein LOC105632434 [Jatropha curcas] |
| 14 |
Hb_006455_020 |
0.1250122015 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_020831_070 |
0.1280993169 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 16 |
Hb_093388_010 |
0.1320141193 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 17 |
Hb_000346_050 |
0.1344812016 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 18 |
Hb_000644_020 |
0.1400877007 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 19 |
Hb_001856_280 |
0.140652322 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 20 |
Hb_001542_010 |
0.1464028671 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |