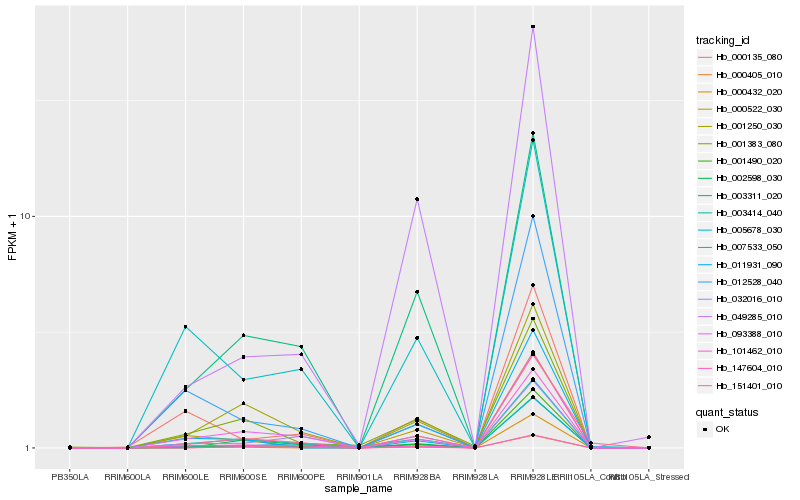
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000432_020 |
0.0 |
desease resistance |
Gene Name: Tropomyosin_1 |
PREDICTED: putative disease resistance protein RGA1 isoform X2 [Jatropha curcas] |
| 2 |
Hb_007533_050 |
0.0950210409 |
- |
- |
- |
| 3 |
Hb_003414_040 |
0.0968536576 |
- |
- |
hypothetical protein VITISV_016691 [Vitis vinifera] |
| 4 |
Hb_003311_020 |
0.1123142336 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_093388_010 |
0.1141093793 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 6 |
Hb_151401_010 |
0.1172629523 |
- |
- |
- |
| 7 |
Hb_001250_030 |
0.1175559249 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000405_010 |
0.1184012413 |
- |
- |
hypothetical protein JCGZ_15779 [Jatropha curcas] |
| 9 |
Hb_000522_030 |
0.1187565962 |
- |
- |
hypothetical protein VITISV_006017 [Vitis vinifera] |
| 10 |
Hb_001490_020 |
0.1313335299 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 11 |
Hb_011931_090 |
0.1314988801 |
- |
- |
- |
| 12 |
Hb_001383_080 |
0.1317036016 |
- |
- |
OSJNBa0019D11.8 [Oryza sativa Japonica Group] |
| 13 |
Hb_049285_010 |
0.1332500055 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 14 |
Hb_005678_030 |
0.133745131 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 15 |
Hb_147604_010 |
0.1360726006 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 16 |
Hb_002598_030 |
0.1402557977 |
- |
- |
- |
| 17 |
Hb_012528_040 |
0.1428671511 |
- |
- |
- |
| 18 |
Hb_032016_010 |
0.1434878788 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At3g28040-like isoform X1 [Citrus sinensis] |
| 19 |
Hb_101462_010 |
0.1478286473 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39030 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000135_080 |
0.1487655823 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |