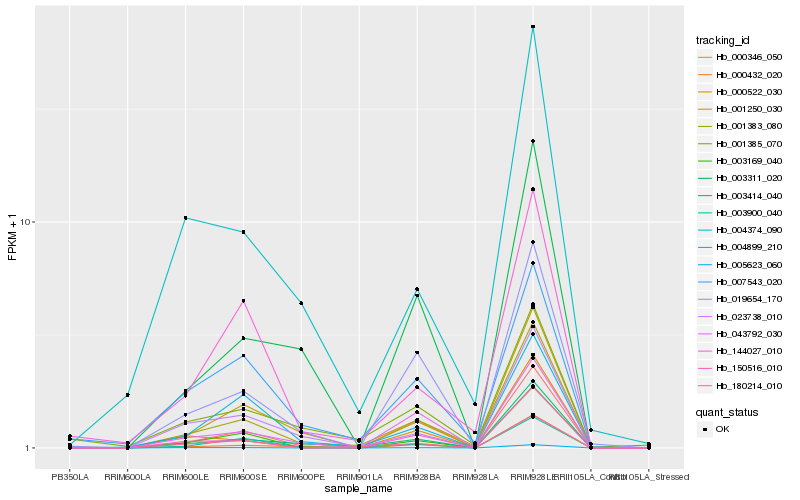
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001383_080 |
0.0 |
- |
- |
OSJNBa0019D11.8 [Oryza sativa Japonica Group] |
| 2 |
Hb_003414_040 |
0.1121704456 |
- |
- |
hypothetical protein VITISV_016691 [Vitis vinifera] |
| 3 |
Hb_001250_030 |
0.1268809033 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_019654_170 |
0.129133062 |
transcription factor |
TF Family: ARID |
PREDICTED: high mobility group B protein 9 [Jatropha curcas] |
| 5 |
Hb_000432_020 |
0.1317036016 |
desease resistance |
Gene Name: Tropomyosin_1 |
PREDICTED: putative disease resistance protein RGA1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000346_050 |
0.133166388 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 7 |
Hb_003900_040 |
0.1351893483 |
- |
- |
hypothetical protein [Lotus japonicus] |
| 8 |
Hb_150516_010 |
0.1372050883 |
- |
- |
PREDICTED: uncharacterized protein LOC104886268, partial [Beta vulgaris subsp. vulgaris] |
| 9 |
Hb_003169_040 |
0.1381662001 |
- |
- |
- |
| 10 |
Hb_180214_010 |
0.1383249299 |
- |
- |
PREDICTED: glutamic acid-rich protein-like [Jatropha curcas] |
| 11 |
Hb_005623_060 |
0.139048564 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 12 |
Hb_144027_010 |
0.1400106755 |
- |
- |
- |
| 13 |
Hb_000522_030 |
0.1429184447 |
- |
- |
hypothetical protein VITISV_006017 [Vitis vinifera] |
| 14 |
Hb_023738_010 |
0.1437368905 |
- |
- |
Concanavalin A-like lectin protein kinase family protein [Theobroma cacao] |
| 15 |
Hb_001385_070 |
0.1452585643 |
- |
- |
hypothetical protein VITISV_019194 [Vitis vinifera] |
| 16 |
Hb_003311_020 |
0.1520268317 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_007543_020 |
0.1536973286 |
- |
- |
PREDICTED: uncharacterized protein LOC104229533, partial [Nicotiana sylvestris] |
| 18 |
Hb_004374_090 |
0.1538516362 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein BEL1 homolog [Populus euphratica] |
| 19 |
Hb_043792_030 |
0.154482347 |
- |
- |
PREDICTED: uncharacterized protein LOC105632434 [Jatropha curcas] |
| 20 |
Hb_004899_210 |
0.158632944 |
- |
- |
PREDICTED: respiratory burst oxidase homolog protein E [Jatropha curcas] |