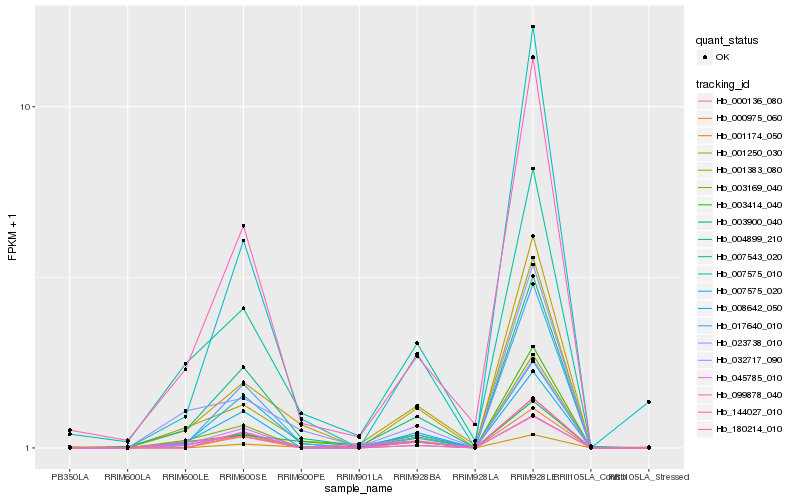
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003900_040 |
0.0 |
- |
- |
hypothetical protein [Lotus japonicus] |
| 2 |
Hb_004899_210 |
0.0732014548 |
- |
- |
PREDICTED: respiratory burst oxidase homolog protein E [Jatropha curcas] |
| 3 |
Hb_144027_010 |
0.1197000273 |
- |
- |
- |
| 4 |
Hb_180214_010 |
0.122758186 |
- |
- |
PREDICTED: glutamic acid-rich protein-like [Jatropha curcas] |
| 5 |
Hb_032717_090 |
0.1264102885 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001250_030 |
0.1322061218 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003169_040 |
0.1333743672 |
- |
- |
- |
| 8 |
Hb_001383_080 |
0.1351893483 |
- |
- |
OSJNBa0019D11.8 [Oryza sativa Japonica Group] |
| 9 |
Hb_017640_010 |
0.1359587581 |
- |
- |
hypothetical protein JCGZ_18815 [Jatropha curcas] |
| 10 |
Hb_008642_050 |
0.1384349887 |
- |
- |
- |
| 11 |
Hb_007575_020 |
0.1556149588 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 12 |
Hb_045785_010 |
0.1580272224 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 13 |
Hb_023738_010 |
0.1593209536 |
- |
- |
Concanavalin A-like lectin protein kinase family protein [Theobroma cacao] |
| 14 |
Hb_000136_080 |
0.1630192759 |
- |
- |
hypothetical protein AALP_AA8G499800 [Arabis alpina] |
| 15 |
Hb_007575_010 |
0.16302485 |
- |
- |
PREDICTED: cytochrome P450 71A1 [Vitis vinifera] |
| 16 |
Hb_007543_020 |
0.1636210857 |
- |
- |
PREDICTED: uncharacterized protein LOC104229533, partial [Nicotiana sylvestris] |
| 17 |
Hb_003414_040 |
0.1689859096 |
- |
- |
hypothetical protein VITISV_016691 [Vitis vinifera] |
| 18 |
Hb_099878_040 |
0.1707966149 |
- |
- |
PREDICTED: uncharacterized protein LOC102703670 [Oryza brachyantha] |
| 19 |
Hb_000975_060 |
0.1728646363 |
- |
- |
- |
| 20 |
Hb_001174_050 |
0.1733386034 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |