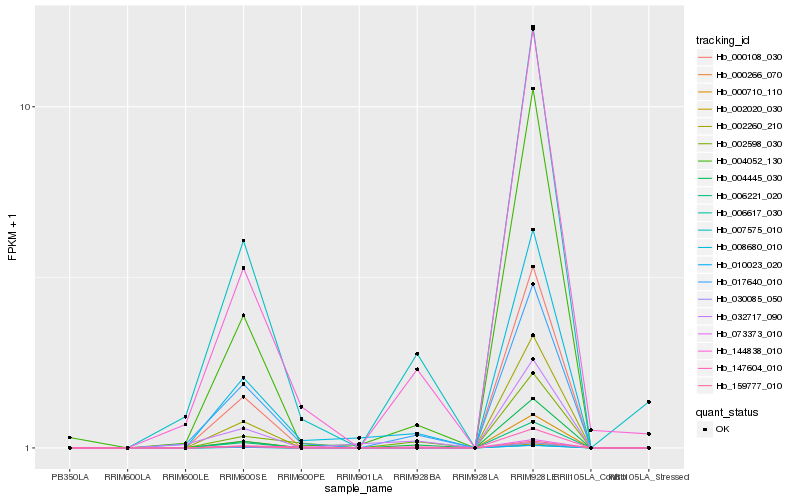
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008680_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_004052_130 |
0.1249340476 |
- |
- |
PREDICTED: cytochrome P450 78A5 [Jatropha curcas] |
| 3 |
Hb_017640_010 |
0.1315493737 |
- |
- |
hypothetical protein JCGZ_18815 [Jatropha curcas] |
| 4 |
Hb_004445_030 |
0.1418480461 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_144838_010 |
0.1420653649 |
- |
- |
hypothetical protein JCGZ_08246 [Jatropha curcas] |
| 6 |
Hb_002598_030 |
0.1442847559 |
- |
- |
- |
| 7 |
Hb_032717_090 |
0.1451826117 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_147604_010 |
0.1545434624 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 9 |
Hb_007575_010 |
0.1572058748 |
- |
- |
PREDICTED: cytochrome P450 71A1 [Vitis vinifera] |
| 10 |
Hb_000710_110 |
0.1648784175 |
- |
- |
Vps51/Vps67 [Citrus endogenous pararetrovirus] |
| 11 |
Hb_000108_030 |
0.1650184197 |
- |
- |
- |
| 12 |
Hb_002260_210 |
0.1650196098 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_006221_020 |
0.1650196152 |
- |
- |
PREDICTED: uncharacterized protein LOC104227769, partial [Nicotiana sylvestris] |
| 14 |
Hb_010023_020 |
0.1671620762 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 15 |
Hb_006617_030 |
0.1671841974 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 16 |
Hb_002020_030 |
0.1673591866 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 17 |
Hb_030085_050 |
0.167529365 |
- |
- |
PREDICTED: uncharacterized protein LOC105797448 [Gossypium raimondii] |
| 18 |
Hb_000266_070 |
0.1675857256 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: uncharacterized protein LOC105765578 [Gossypium raimondii] |
| 19 |
Hb_159777_010 |
0.1678149362 |
- |
- |
JHL22C18.10 [Jatropha curcas] |
| 20 |
Hb_073373_010 |
0.1680813995 |
- |
- |
hypothetical protein JCGZ_11284 [Jatropha curcas] |