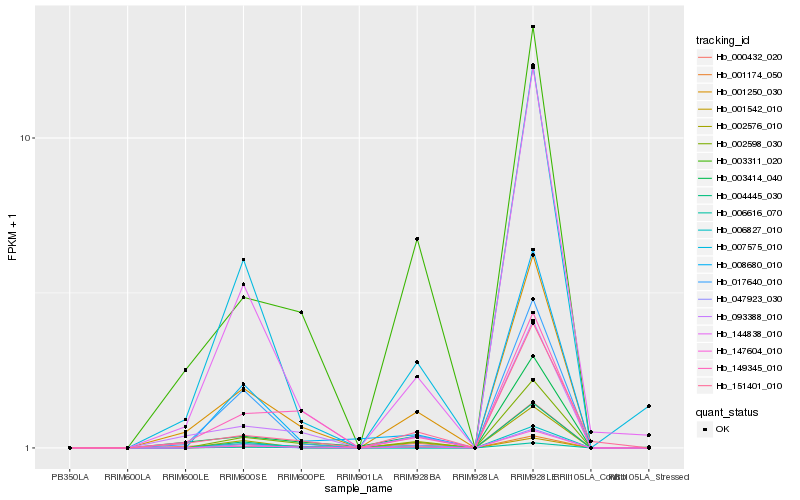
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002598_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_147604_010 |
0.0339394858 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 3 |
Hb_001542_010 |
0.075707678 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 4 |
Hb_144838_010 |
0.1257546091 |
- |
- |
hypothetical protein JCGZ_08246 [Jatropha curcas] |
| 5 |
Hb_001250_030 |
0.1333798548 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000432_020 |
0.1402557977 |
desease resistance |
Gene Name: Tropomyosin_1 |
PREDICTED: putative disease resistance protein RGA1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_008680_010 |
0.1442847559 |
- |
- |
- |
| 8 |
Hb_004445_030 |
0.1459947381 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_007575_010 |
0.1580948578 |
- |
- |
PREDICTED: cytochrome P450 71A1 [Vitis vinifera] |
| 10 |
Hb_093388_010 |
0.1584567956 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 11 |
Hb_017640_010 |
0.1592109891 |
- |
- |
hypothetical protein JCGZ_18815 [Jatropha curcas] |
| 12 |
Hb_001174_050 |
0.1598303934 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 13 |
Hb_006616_070 |
0.1648216951 |
- |
- |
PREDICTED: uncharacterized protein LOC104894399 [Beta vulgaris subsp. vulgaris] |
| 14 |
Hb_003414_040 |
0.1656895152 |
- |
- |
hypothetical protein VITISV_016691 [Vitis vinifera] |
| 15 |
Hb_047923_030 |
0.1660385288 |
- |
- |
hypothetical protein JCGZ_09181 [Jatropha curcas] |
| 16 |
Hb_002576_010 |
0.1668885987 |
- |
- |
- |
| 17 |
Hb_003311_020 |
0.1689383605 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_151401_010 |
0.1718045373 |
- |
- |
- |
| 19 |
Hb_006827_010 |
0.1740800342 |
- |
- |
retrotransposon protein, putative, unclassified, expressed [Oryza sativa Japonica Group] |
| 20 |
Hb_149345_010 |
0.1743874164 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |