| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
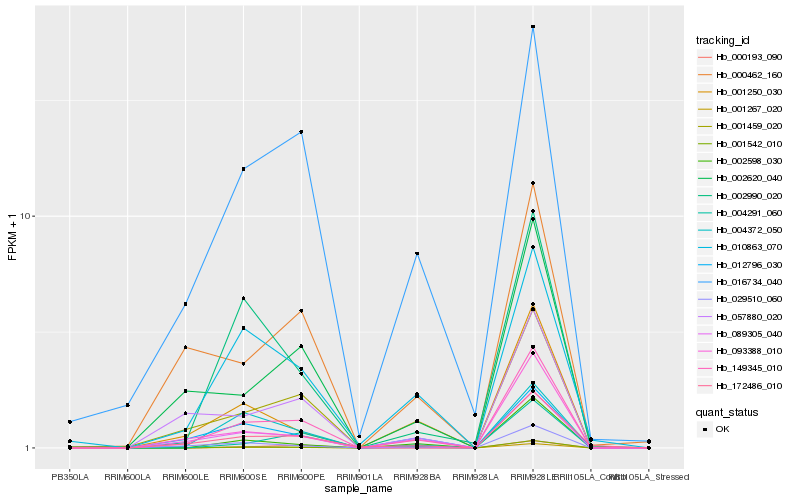
Hb_149345_010 |
0.0 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 2 |
Hb_001459_020 |
0.125416966 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 3 |
Hb_093388_010 |
0.1321393676 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 4 |
Hb_016734_040 |
0.1389259489 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 5 |
Hb_029510_060 |
0.1397367699 |
- |
- |
PREDICTED: probable plastidic glucose transporter 3 [Jatropha curcas] |
| 6 |
Hb_001542_010 |
0.1399031293 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 7 |
Hb_172486_010 |
0.1400473369 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 8 |
Hb_002620_040 |
0.1418659736 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 9 |
Hb_000193_090 |
0.1484304796 |
- |
- |
cysteine protease, putative [Ricinus communis] |
| 10 |
Hb_057880_020 |
0.1537486011 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1, partial [Jatropha curcas] |
| 11 |
Hb_001250_030 |
0.1576406467 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004291_060 |
0.1607367618 |
- |
- |
hypothetical protein F775_42457 [Aegilops tauschii] |
| 13 |
Hb_010863_070 |
0.1615423613 |
- |
- |
PREDICTED: glutaredoxin-C11 [Jatropha curcas] |
| 14 |
Hb_089305_040 |
0.1650870556 |
- |
- |
- |
| 15 |
Hb_000462_160 |
0.1675170373 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 16 |
Hb_012796_030 |
0.168008017 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 17 |
Hb_004372_050 |
0.1686161236 |
- |
- |
putative receptor-like protein kinase [Morus notabilis] |
| 18 |
Hb_001267_020 |
0.1705283949 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 19 |
Hb_002990_020 |
0.1706201931 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 20 |
Hb_002598_030 |
0.1743874164 |
- |
- |
- |