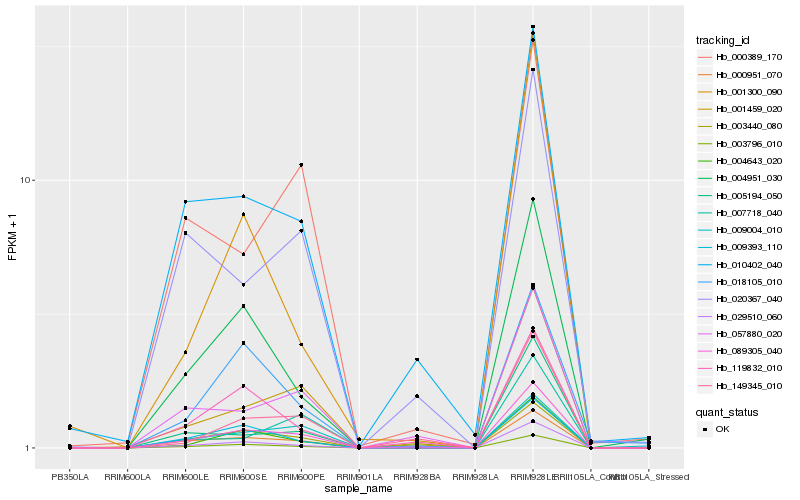
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_089305_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_003796_010 |
0.0638999204 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 3 |
Hb_000951_070 |
0.1010113311 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: receptor-like protein kinase FERONIA [Populus euphratica] |
| 4 |
Hb_004643_020 |
0.1032174063 |
- |
- |
hypothetical protein AMTR_s05716p00004100, partial [Amborella trichopoda] |
| 5 |
Hb_001459_020 |
0.1124383829 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 6 |
Hb_009393_110 |
0.1316910343 |
- |
- |
PREDICTED: circadian locomoter output cycles protein kaput [Jatropha curcas] |
| 7 |
Hb_004951_030 |
0.132476448 |
- |
- |
9-cis-epoxycarotenoid dioxygenase, putative [Ricinus communis] |
| 8 |
Hb_029510_060 |
0.1415457988 |
- |
- |
PREDICTED: probable plastidic glucose transporter 3 [Jatropha curcas] |
| 9 |
Hb_057880_020 |
0.1520384676 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1, partial [Jatropha curcas] |
| 10 |
Hb_005194_050 |
0.1549086371 |
transcription factor |
TF Family: GRAS |
SCL domain class transcription factor [Theobroma cacao] |
| 11 |
Hb_119832_010 |
0.1555958121 |
- |
- |
- |
| 12 |
Hb_010402_040 |
0.1561093422 |
- |
- |
hypothetical protein JCGZ_21227 [Jatropha curcas] |
| 13 |
Hb_009004_010 |
0.1601287191 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 14 |
Hb_001300_090 |
0.1637364492 |
- |
- |
PREDICTED: glutaredoxin-C11 [Jatropha curcas] |
| 15 |
Hb_149345_010 |
0.1650870556 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 16 |
Hb_000389_170 |
0.1686997306 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 17 |
Hb_018105_010 |
0.1696453769 |
- |
- |
hypothetical protein POPTR_0004s08710g [Populus trichocarpa] |
| 18 |
Hb_007718_040 |
0.1700890138 |
- |
- |
hypothetical protein PRUPE_ppb025054mg [Prunus persica] |
| 19 |
Hb_003440_080 |
0.1720855631 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 20 |
Hb_020367_040 |
0.1736007923 |
- |
- |
hypothetical protein JCGZ_01061 [Jatropha curcas] |