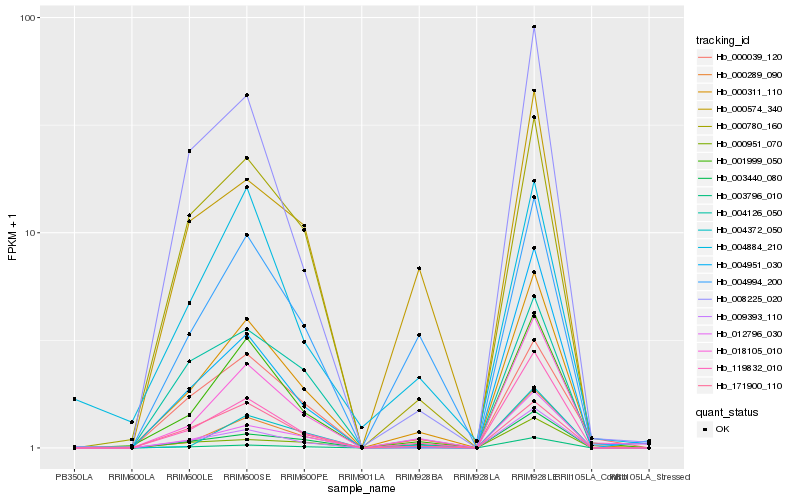
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000311_110 |
0.0 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 2 |
Hb_001999_050 |
0.0793307907 |
rubber biosynthesis |
Gene Name: CPT7 |
PREDICTED: dehydrodolichyl diphosphate synthase 2 [Jatropha curcas] |
| 3 |
Hb_119832_010 |
0.0877359876 |
- |
- |
- |
| 4 |
Hb_018105_010 |
0.1180243038 |
- |
- |
hypothetical protein POPTR_0004s08710g [Populus trichocarpa] |
| 5 |
Hb_009393_110 |
0.1215522247 |
- |
- |
PREDICTED: circadian locomoter output cycles protein kaput [Jatropha curcas] |
| 6 |
Hb_003440_080 |
0.1230148245 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 7 |
Hb_000780_160 |
0.1243186224 |
- |
- |
hypothetical protein RCOM_0483300 [Ricinus communis] |
| 8 |
Hb_000289_090 |
0.1365613138 |
- |
- |
- |
| 9 |
Hb_004994_200 |
0.1378850436 |
- |
- |
PREDICTED: uncharacterized protein LOC105174340 [Sesamum indicum] |
| 10 |
Hb_008225_020 |
0.1404384062 |
- |
- |
PREDICTED: cytochrome P450 71A26-like [Jatropha curcas] |
| 11 |
Hb_000039_120 |
0.1433358432 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 3-like [Jatropha curcas] |
| 12 |
Hb_171900_110 |
0.1457297463 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT3-like [Populus euphratica] |
| 13 |
Hb_000951_070 |
0.1479174259 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: receptor-like protein kinase FERONIA [Populus euphratica] |
| 14 |
Hb_012796_030 |
0.1544925267 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 15 |
Hb_000574_340 |
0.1558389327 |
- |
- |
hypothetical protein RCOM_0901420 [Ricinus communis] |
| 16 |
Hb_004951_030 |
0.1584828753 |
- |
- |
9-cis-epoxycarotenoid dioxygenase, putative [Ricinus communis] |
| 17 |
Hb_003796_010 |
0.1625048543 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 18 |
Hb_004884_210 |
0.1648059982 |
- |
- |
PREDICTED: mitoferrin-like [Jatropha curcas] |
| 19 |
Hb_004372_050 |
0.1662099023 |
- |
- |
putative receptor-like protein kinase [Morus notabilis] |
| 20 |
Hb_004126_050 |
0.1683547789 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VII.1 [Tarenaya hassleriana] |