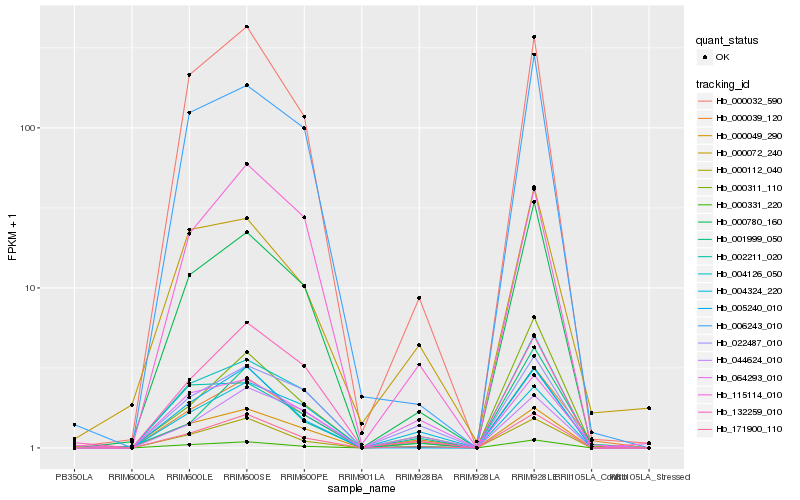
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000039_120 |
0.0 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 3-like [Jatropha curcas] |
| 2 |
Hb_171900_110 |
0.1134105578 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT3-like [Populus euphratica] |
| 3 |
Hb_000780_160 |
0.1165450904 |
- |
- |
hypothetical protein RCOM_0483300 [Ricinus communis] |
| 4 |
Hb_000032_590 |
0.1401182217 |
- |
- |
PREDICTED: glutaredoxin domain-containing cysteine-rich protein 1 [Jatropha curcas] |
| 5 |
Hb_000311_110 |
0.1433358432 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 6 |
Hb_001999_050 |
0.143439546 |
rubber biosynthesis |
Gene Name: CPT7 |
PREDICTED: dehydrodolichyl diphosphate synthase 2 [Jatropha curcas] |
| 7 |
Hb_004126_050 |
0.1449047533 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VII.1 [Tarenaya hassleriana] |
| 8 |
Hb_022487_010 |
0.1455116585 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 9 |
Hb_006243_010 |
0.1464282388 |
- |
- |
PREDICTED: cell wall protein IFF6 [Populus euphratica] |
| 10 |
Hb_000112_040 |
0.1466792942 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 11 |
Hb_005240_010 |
0.153678295 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-B2 [Jatropha curcas] |
| 12 |
Hb_000072_240 |
0.1598613827 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger protein, putative [Ricinus communis] |
| 13 |
Hb_002211_020 |
0.1602695029 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 5-like [Jatropha curcas] |
| 14 |
Hb_000049_290 |
0.1603563892 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1 [Jatropha curcas] |
| 15 |
Hb_132259_010 |
0.1641612237 |
- |
- |
PREDICTED: WAT1-related protein At5g64700 [Jatropha curcas] |
| 16 |
Hb_000331_220 |
0.165668659 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_115114_010 |
0.1665542938 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004324_220 |
0.1677954025 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 53 [Jatropha curcas] |
| 19 |
Hb_064293_010 |
0.1703285435 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 20 |
Hb_044624_010 |
0.1717732102 |
- |
- |
- |