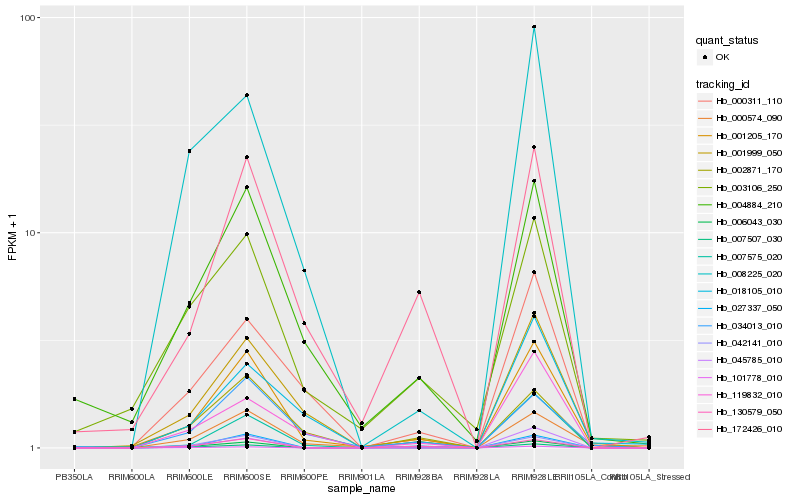
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001205_170 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633940 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001999_050 |
0.1280427046 |
rubber biosynthesis |
Gene Name: CPT7 |
PREDICTED: dehydrodolichyl diphosphate synthase 2 [Jatropha curcas] |
| 3 |
Hb_004884_210 |
0.1500790459 |
- |
- |
PREDICTED: mitoferrin-like [Jatropha curcas] |
| 4 |
Hb_007575_020 |
0.1585201759 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 5 |
Hb_172426_010 |
0.1585836939 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Jatropha curcas] |
| 6 |
Hb_027337_050 |
0.1637242712 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 7 |
Hb_034013_010 |
0.1669021309 |
- |
- |
hypothetical protein JCGZ_21944 [Jatropha curcas] |
| 8 |
Hb_000311_110 |
0.1703090552 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 9 |
Hb_007507_030 |
0.1799641282 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 10 |
Hb_006043_030 |
0.1811932241 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 11 |
Hb_042141_010 |
0.1822209387 |
- |
- |
- |
| 12 |
Hb_000574_090 |
0.1833996063 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 13 |
Hb_130579_050 |
0.1834131523 |
- |
- |
hypothetical protein JCGZ_25577 [Jatropha curcas] |
| 14 |
Hb_003106_250 |
0.1841640833 |
- |
- |
hypothetical protein POPTR_0008s13520g [Populus trichocarpa] |
| 15 |
Hb_002871_170 |
0.1857335485 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein 21/22, putative [Ricinus communis] |
| 16 |
Hb_045785_010 |
0.1874136456 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 17 |
Hb_101778_010 |
0.1881530422 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 18 |
Hb_119832_010 |
0.1909604798 |
- |
- |
- |
| 19 |
Hb_008225_020 |
0.1969486429 |
- |
- |
PREDICTED: cytochrome P450 71A26-like [Jatropha curcas] |
| 20 |
Hb_018105_010 |
0.2006886473 |
- |
- |
hypothetical protein POPTR_0004s08710g [Populus trichocarpa] |