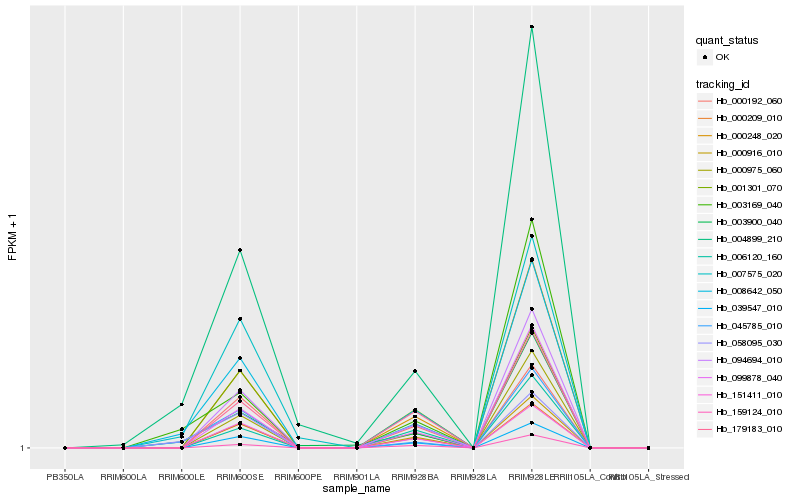
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008642_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_045785_010 |
0.0989484715 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 3 |
Hb_004899_210 |
0.1284090877 |
- |
- |
PREDICTED: respiratory burst oxidase homolog protein E [Jatropha curcas] |
| 4 |
Hb_058095_030 |
0.1343924604 |
- |
- |
- |
| 5 |
Hb_094694_010 |
0.1363983667 |
- |
- |
- |
| 6 |
Hb_039547_010 |
0.136667809 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 3-like [Jatropha curcas] |
| 7 |
Hb_000916_010 |
0.1368280664 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 8 |
Hb_003900_040 |
0.1384349887 |
- |
- |
hypothetical protein [Lotus japonicus] |
| 9 |
Hb_000248_020 |
0.1391583185 |
- |
- |
PREDICTED: uncharacterized protein LOC104243878 [Nicotiana sylvestris] |
| 10 |
Hb_003169_040 |
0.1402383489 |
- |
- |
- |
| 11 |
Hb_007575_020 |
0.1409615887 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 12 |
Hb_000209_010 |
0.142354461 |
- |
- |
- |
| 13 |
Hb_000975_060 |
0.1451034739 |
- |
- |
- |
| 14 |
Hb_151411_010 |
0.1471672122 |
- |
- |
- |
| 15 |
Hb_000192_060 |
0.1529950426 |
- |
- |
- |
| 16 |
Hb_099878_040 |
0.1534801789 |
- |
- |
PREDICTED: uncharacterized protein LOC102703670 [Oryza brachyantha] |
| 17 |
Hb_179183_010 |
0.1594884238 |
- |
- |
RALF-LIKE 32 family protein [Populus trichocarpa] |
| 18 |
Hb_159124_010 |
0.1595219258 |
- |
- |
PREDICTED: uncharacterized protein LOC103332224 [Prunus mume] |
| 19 |
Hb_001301_070 |
0.1626809154 |
- |
- |
- |
| 20 |
Hb_006120_160 |
0.1704008736 |
- |
- |
- |