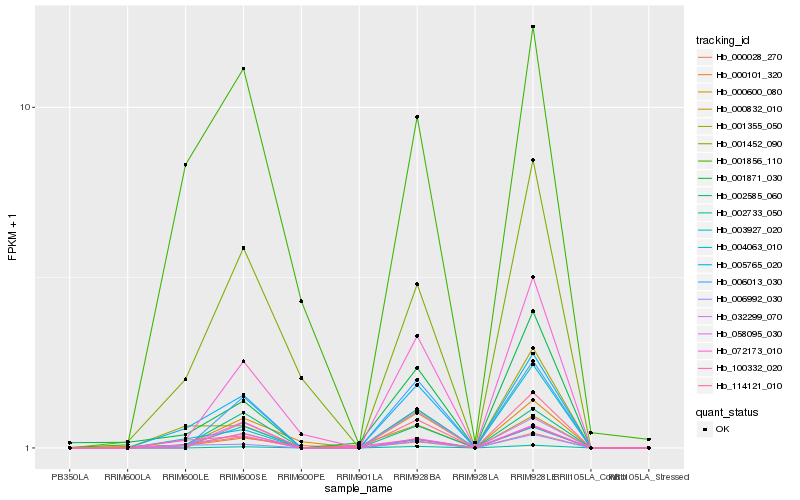
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005765_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_006992_030 |
0.1020409107 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 3 |
Hb_058095_030 |
0.1029546732 |
- |
- |
- |
| 4 |
Hb_000028_270 |
0.1251635971 |
- |
- |
- |
| 5 |
Hb_002585_060 |
0.1509377915 |
- |
- |
PREDICTED: uncharacterized protein LOC104899128, partial [Beta vulgaris subsp. vulgaris] |
| 6 |
Hb_003927_020 |
0.1547048085 |
- |
- |
PREDICTED: uncharacterized protein LOC105782546 [Gossypium raimondii] |
| 7 |
Hb_032299_070 |
0.1683623973 |
- |
- |
- |
| 8 |
Hb_072173_010 |
0.1710476911 |
- |
- |
PREDICTED: putative cysteine-rich receptor-like protein kinase 34 [Jatropha curcas] |
| 9 |
Hb_001355_050 |
0.1758739637 |
- |
- |
- |
| 10 |
Hb_000101_320 |
0.1788051921 |
- |
- |
- |
| 11 |
Hb_000832_010 |
0.1854223853 |
- |
- |
hypothetical protein VITISV_005044 [Vitis vinifera] |
| 12 |
Hb_000600_080 |
0.1854322174 |
- |
- |
- |
| 13 |
Hb_001871_030 |
0.1860871295 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_100332_020 |
0.187662876 |
- |
- |
PREDICTED: uncharacterized protein LOC105168695 [Sesamum indicum] |
| 15 |
Hb_002733_050 |
0.1911922584 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 16 |
Hb_004063_010 |
0.1923460631 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_006013_030 |
0.1935932974 |
- |
- |
- |
| 18 |
Hb_001452_090 |
0.194181984 |
- |
- |
PREDICTED: serine carboxypeptidase II-3-like [Vitis vinifera] |
| 19 |
Hb_114121_010 |
0.195783939 |
- |
- |
- |
| 20 |
Hb_001856_110 |
0.1962253777 |
- |
- |
PREDICTED: cellulose synthase-like protein G2 [Populus euphratica] |