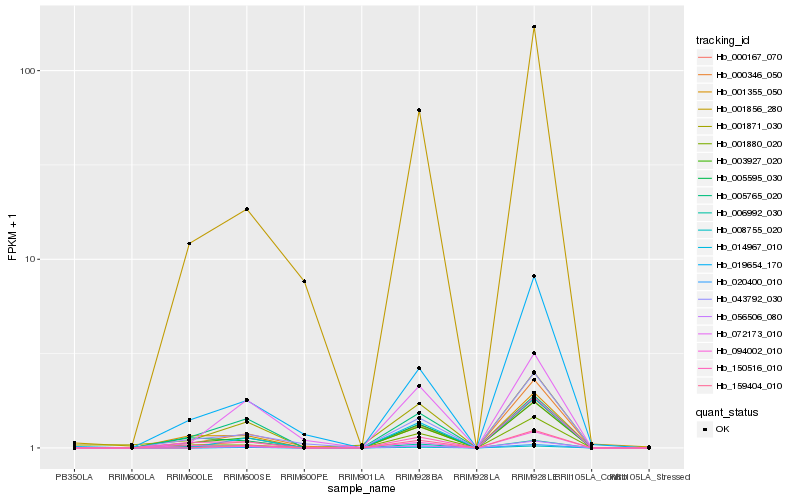
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003927_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105782546 [Gossypium raimondii] |
| 2 |
Hb_000167_070 |
0.0877623144 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 3 |
Hb_001880_020 |
0.089515404 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 4 |
Hb_001355_050 |
0.0913705305 |
- |
- |
- |
| 5 |
Hb_005595_030 |
0.0929043152 |
- |
- |
hypothetical protein EUGRSUZ_A00624 [Eucalyptus grandis] |
| 6 |
Hb_006992_030 |
0.1002682103 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 7 |
Hb_000346_050 |
0.1157467763 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 8 |
Hb_043792_030 |
0.1240291704 |
- |
- |
PREDICTED: uncharacterized protein LOC105632434 [Jatropha curcas] |
| 9 |
Hb_001856_280 |
0.1301600246 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 10 |
Hb_019654_170 |
0.1478537045 |
transcription factor |
TF Family: ARID |
PREDICTED: high mobility group B protein 9 [Jatropha curcas] |
| 11 |
Hb_150516_010 |
0.1494823233 |
- |
- |
PREDICTED: uncharacterized protein LOC104886268, partial [Beta vulgaris subsp. vulgaris] |
| 12 |
Hb_001871_030 |
0.1531479 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005765_020 |
0.1547048085 |
- |
- |
- |
| 14 |
Hb_094002_010 |
0.1590416465 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Vitis vinifera] |
| 15 |
Hb_014967_010 |
0.1630986282 |
- |
- |
- |
| 16 |
Hb_159404_010 |
0.1636525258 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 17 |
Hb_008755_020 |
0.1716846465 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_020400_010 |
0.1748558895 |
- |
- |
PREDICTED: uncharacterized protein LOC105644073 [Jatropha curcas] |
| 19 |
Hb_056506_080 |
0.1758328128 |
- |
- |
PREDICTED: uncharacterized protein LOC104888944 [Beta vulgaris subsp. vulgaris] |
| 20 |
Hb_072173_010 |
0.1779248679 |
- |
- |
PREDICTED: putative cysteine-rich receptor-like protein kinase 34 [Jatropha curcas] |