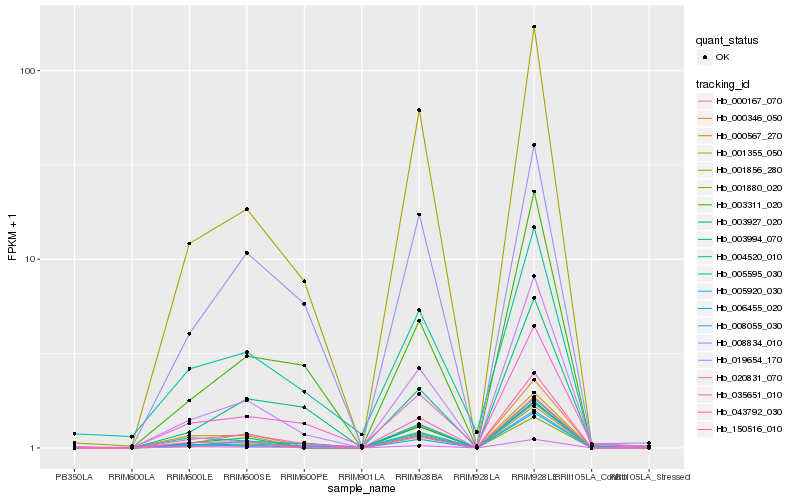
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_043792_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632434 [Jatropha curcas] |
| 2 |
Hb_001856_280 |
0.0455794033 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 3 |
Hb_000346_050 |
0.0680997428 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 4 |
Hb_019654_170 |
0.0704628471 |
transcription factor |
TF Family: ARID |
PREDICTED: high mobility group B protein 9 [Jatropha curcas] |
| 5 |
Hb_020831_070 |
0.092500972 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 6 |
Hb_005920_030 |
0.1060649514 |
- |
- |
putative polyprotein [Oryza sativa Japonica Group] |
| 7 |
Hb_003311_020 |
0.122291701 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_006455_020 |
0.1228639145 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_003927_020 |
0.1240291704 |
- |
- |
PREDICTED: uncharacterized protein LOC105782546 [Gossypium raimondii] |
| 10 |
Hb_008055_030 |
0.1307708889 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 11 |
Hb_001880_020 |
0.1346668961 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 12 |
Hb_000167_070 |
0.1371909464 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 13 |
Hb_008834_010 |
0.1393676641 |
- |
- |
hypothetical protein POPTR_0016s04660g [Populus trichocarpa] |
| 14 |
Hb_150516_010 |
0.1405728271 |
- |
- |
PREDICTED: uncharacterized protein LOC104886268, partial [Beta vulgaris subsp. vulgaris] |
| 15 |
Hb_035651_010 |
0.1415150815 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 16 |
Hb_001355_050 |
0.141826526 |
- |
- |
- |
| 17 |
Hb_005595_030 |
0.1443321132 |
- |
- |
hypothetical protein EUGRSUZ_A00624 [Eucalyptus grandis] |
| 18 |
Hb_000567_270 |
0.144517141 |
- |
- |
hypothetical protein L484_017162 [Morus notabilis] |
| 19 |
Hb_003994_070 |
0.1478171888 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 20 |
Hb_004520_010 |
0.1478253512 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |