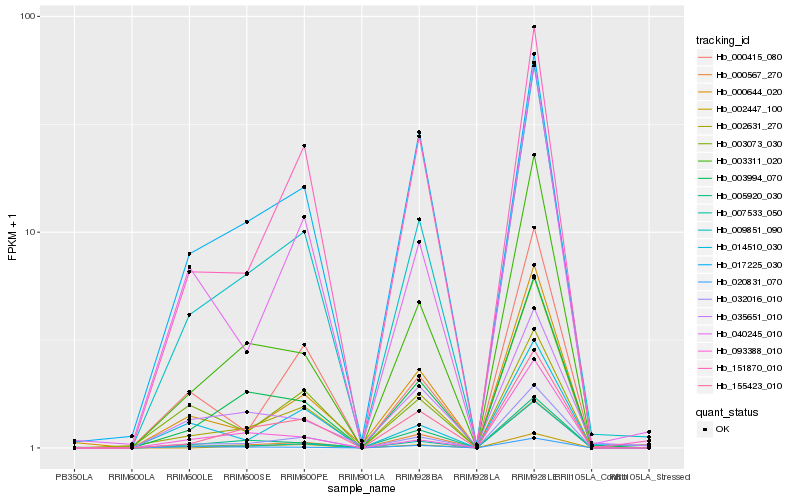
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009851_090 |
0.0 |
- |
- |
Protein ABIL2, putative [Ricinus communis] |
| 2 |
Hb_032016_010 |
0.0815970962 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At3g28040-like isoform X1 [Citrus sinensis] |
| 3 |
Hb_003994_070 |
0.0835562832 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 4 |
Hb_003311_020 |
0.0920416506 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_155423_010 |
0.0987778123 |
- |
- |
- |
| 6 |
Hb_002631_270 |
0.11092406 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0005s09930g [Populus trichocarpa] |
| 7 |
Hb_000567_270 |
0.1175885602 |
- |
- |
hypothetical protein L484_017162 [Morus notabilis] |
| 8 |
Hb_151870_010 |
0.1191452481 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_040245_010 |
0.122128679 |
- |
- |
hypothetical protein CISIN_1g039575mg, partial [Citrus sinensis] |
| 10 |
Hb_000644_020 |
0.1247129358 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 11 |
Hb_003073_030 |
0.1264381618 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_035651_010 |
0.1280851759 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 13 |
Hb_007533_050 |
0.1391882842 |
- |
- |
- |
| 14 |
Hb_000415_080 |
0.1421519082 |
- |
- |
- |
| 15 |
Hb_020831_070 |
0.1448371581 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 16 |
Hb_005920_030 |
0.1462382138 |
- |
- |
putative polyprotein [Oryza sativa Japonica Group] |
| 17 |
Hb_017225_030 |
0.1478138615 |
- |
- |
PREDICTED: probable cytosolic oligopeptidase A [Jatropha curcas] |
| 18 |
Hb_014510_030 |
0.1522022474 |
- |
- |
hypothetical protein VITISV_027576 [Vitis vinifera] |
| 19 |
Hb_002447_100 |
0.1528981617 |
- |
- |
hypothetical protein POPTR_0009s06260g [Populus trichocarpa] |
| 20 |
Hb_093388_010 |
0.1551653474 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |