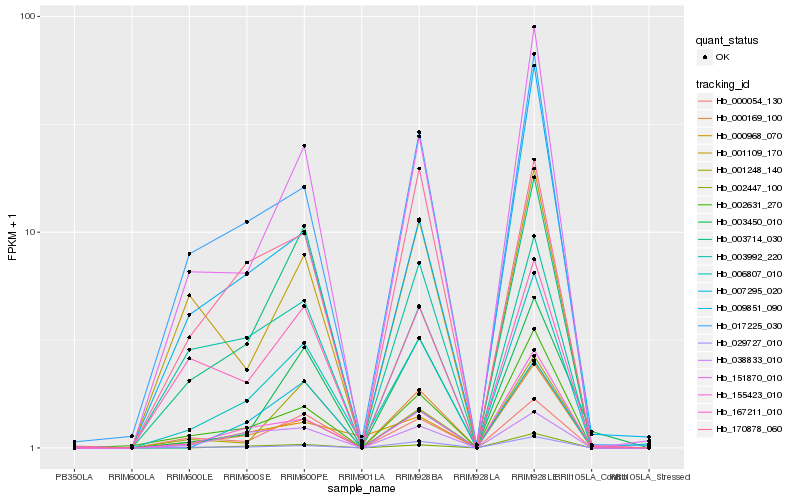
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006807_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_20881 [Jatropha curcas] |
| 2 |
Hb_151870_010 |
0.1051686494 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 3 |
Hb_002631_270 |
0.1192879589 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0005s09930g [Populus trichocarpa] |
| 4 |
Hb_000169_100 |
0.1196338622 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Fragaria vesca subsp. vesca] |
| 5 |
Hb_155423_010 |
0.1243077323 |
- |
- |
- |
| 6 |
Hb_017225_030 |
0.1272554706 |
- |
- |
PREDICTED: probable cytosolic oligopeptidase A [Jatropha curcas] |
| 7 |
Hb_029727_010 |
0.1455762254 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 8 |
Hb_000054_130 |
0.1472219595 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 9 |
Hb_003714_030 |
0.1513679024 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 10 |
Hb_038833_010 |
0.1514292209 |
- |
- |
polyprotein [Ananas comosus] |
| 11 |
Hb_001248_140 |
0.1611839654 |
- |
- |
Thylakoid lumenal 19 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_000968_070 |
0.1613959498 |
- |
- |
- |
| 13 |
Hb_002447_100 |
0.1677862744 |
- |
- |
hypothetical protein POPTR_0009s06260g [Populus trichocarpa] |
| 14 |
Hb_003450_010 |
0.1691132411 |
- |
- |
- |
| 15 |
Hb_167211_010 |
0.1708780641 |
- |
- |
PREDICTED: glutamate receptor 3.4-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_007295_020 |
0.1714384409 |
- |
- |
Uncharacterized protein TCM_034658 [Theobroma cacao] |
| 17 |
Hb_001109_170 |
0.1725240256 |
- |
- |
PREDICTED: urea-proton symporter DUR3 isoform X2 [Jatropha curcas] |
| 18 |
Hb_170878_060 |
0.1753524143 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 19 |
Hb_009851_090 |
0.17752236 |
- |
- |
Protein ABIL2, putative [Ricinus communis] |
| 20 |
Hb_003992_220 |
0.1777564734 |
- |
- |
s-receptor kinase, putative [Ricinus communis] |