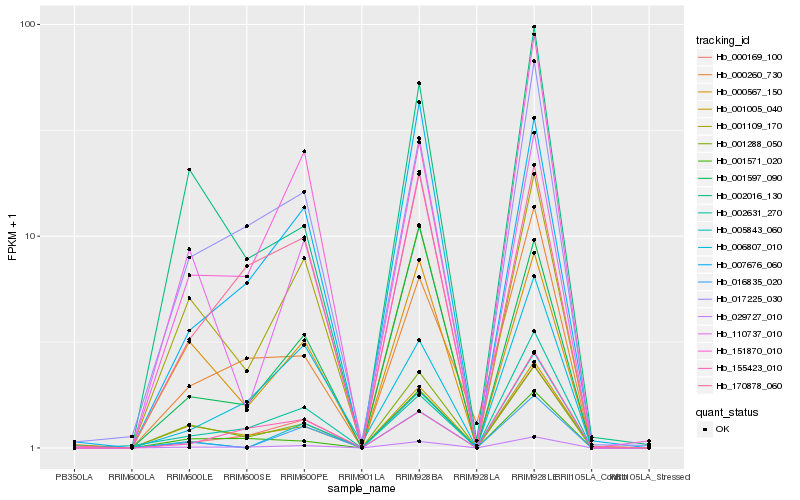
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029727_010 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 2 |
Hb_000169_100 |
0.0486650249 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Fragaria vesca subsp. vesca] |
| 3 |
Hb_001005_040 |
0.0989291269 |
- |
- |
- |
| 4 |
Hb_017225_030 |
0.1135382107 |
- |
- |
PREDICTED: probable cytosolic oligopeptidase A [Jatropha curcas] |
| 5 |
Hb_001288_050 |
0.1247481208 |
- |
- |
- |
| 6 |
Hb_000260_730 |
0.1248432966 |
- |
- |
- |
| 7 |
Hb_002631_270 |
0.128704821 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0005s09930g [Populus trichocarpa] |
| 8 |
Hb_151870_010 |
0.1294561021 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_001597_090 |
0.1416010785 |
- |
- |
- |
| 10 |
Hb_002016_130 |
0.1437796568 |
desease resistance |
Gene Name: ABC_membrane |
multidrug resistance-associated protein 2, 6 (mrp2, 6), abc-transoprter, putative [Ricinus communis] |
| 11 |
Hb_001109_170 |
0.1441883305 |
- |
- |
PREDICTED: urea-proton symporter DUR3 isoform X2 [Jatropha curcas] |
| 12 |
Hb_006807_010 |
0.1455762254 |
- |
- |
hypothetical protein JCGZ_20881 [Jatropha curcas] |
| 13 |
Hb_005843_060 |
0.1474740257 |
- |
- |
PREDICTED: protein FAR-RED IMPAIRED RESPONSE 1 isoform X3 [Nelumbo nucifera] |
| 14 |
Hb_007676_060 |
0.1489953212 |
- |
- |
PREDICTED: cellulose synthase-like protein E2 [Pyrus x bretschneideri] |
| 15 |
Hb_016835_020 |
0.1579283184 |
- |
- |
unnamed protein product [Coffea canephora] |
| 16 |
Hb_155423_010 |
0.1612058431 |
- |
- |
- |
| 17 |
Hb_001571_020 |
0.1646106117 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850-like [Citrus sinensis] |
| 18 |
Hb_000567_150 |
0.1648443382 |
- |
- |
cysteine protease, putative [Ricinus communis] |
| 19 |
Hb_170878_060 |
0.1668045153 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 20 |
Hb_110737_010 |
0.1671382736 |
- |
- |
PREDICTED: aldose 1-epimerase-like [Jatropha curcas] |