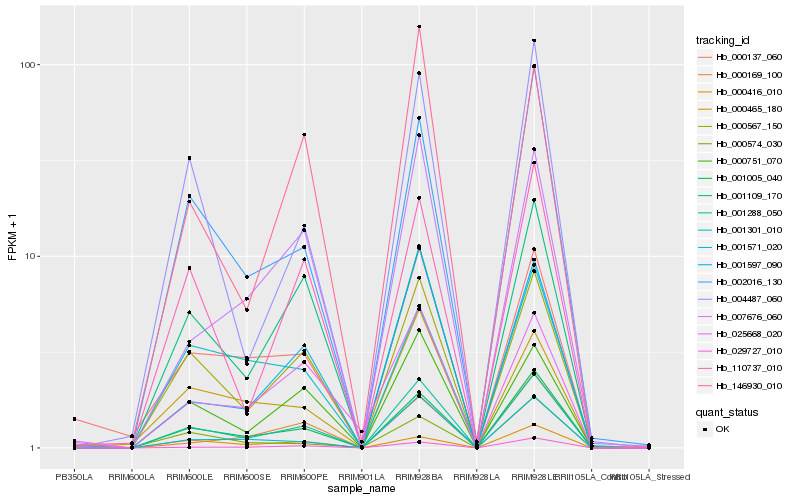
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001288_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_000567_150 |
0.0709396348 |
- |
- |
cysteine protease, putative [Ricinus communis] |
| 3 |
Hb_001005_040 |
0.0746393024 |
- |
- |
- |
| 4 |
Hb_000751_070 |
0.1001102343 |
- |
- |
- |
| 5 |
Hb_001597_090 |
0.1112017689 |
- |
- |
- |
| 6 |
Hb_001571_020 |
0.1141889342 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850-like [Citrus sinensis] |
| 7 |
Hb_002016_130 |
0.117201046 |
desease resistance |
Gene Name: ABC_membrane |
multidrug resistance-associated protein 2, 6 (mrp2, 6), abc-transoprter, putative [Ricinus communis] |
| 8 |
Hb_029727_010 |
0.1247481208 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 9 |
Hb_110737_010 |
0.1282870439 |
- |
- |
PREDICTED: aldose 1-epimerase-like [Jatropha curcas] |
| 10 |
Hb_000137_060 |
0.1283039726 |
- |
- |
hypothetical protein JCGZ_15115 [Jatropha curcas] |
| 11 |
Hb_004487_060 |
0.133625849 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 12 |
Hb_000465_180 |
0.1346833452 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 13 |
Hb_007676_060 |
0.136202701 |
- |
- |
PREDICTED: cellulose synthase-like protein E2 [Pyrus x bretschneideri] |
| 14 |
Hb_001109_170 |
0.1391558345 |
- |
- |
PREDICTED: urea-proton symporter DUR3 isoform X2 [Jatropha curcas] |
| 15 |
Hb_146930_010 |
0.1457494196 |
- |
- |
- |
| 16 |
Hb_000169_100 |
0.1528339134 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Fragaria vesca subsp. vesca] |
| 17 |
Hb_025668_020 |
0.1539219687 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000574_030 |
0.159446593 |
desease resistance |
Gene Name: Glyco_hydro_38 |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 19 |
Hb_000416_010 |
0.1640817263 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 20 |
Hb_001301_010 |
0.1641270469 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |