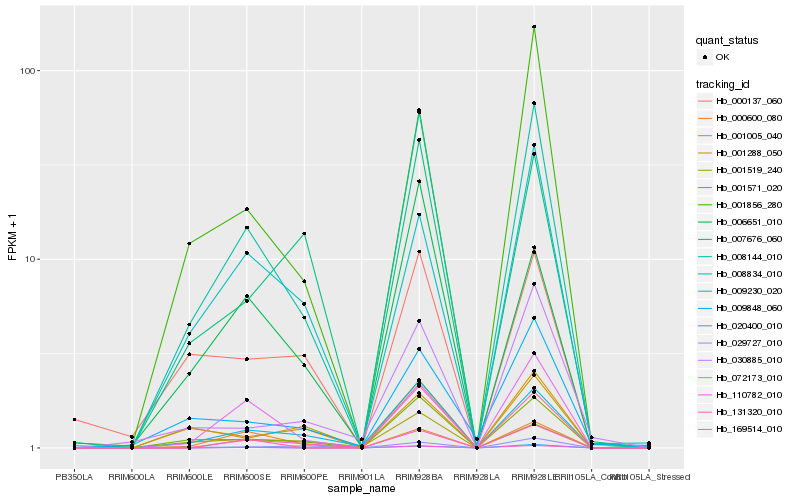
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008144_010 |
0.0 |
- |
- |
hypothetical protein VITISV_015004 [Vitis vinifera] |
| 2 |
Hb_001571_020 |
0.0964031793 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850-like [Citrus sinensis] |
| 3 |
Hb_009230_020 |
0.1396735012 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_072173_010 |
0.1565424895 |
- |
- |
PREDICTED: putative cysteine-rich receptor-like protein kinase 34 [Jatropha curcas] |
| 5 |
Hb_131320_010 |
0.1617601272 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 isoform X2 [Jatropha curcas] |
| 6 |
Hb_110782_010 |
0.1659606925 |
- |
- |
PREDICTED: uncharacterized protein LOC105797448 [Gossypium raimondii] |
| 7 |
Hb_009848_060 |
0.177917357 |
- |
- |
unknown [Glycine max] |
| 8 |
Hb_008834_010 |
0.1812803863 |
- |
- |
hypothetical protein POPTR_0016s04660g [Populus trichocarpa] |
| 9 |
Hb_001856_280 |
0.1825219353 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 10 |
Hb_000600_080 |
0.183885482 |
- |
- |
- |
| 11 |
Hb_020400_010 |
0.1867090158 |
- |
- |
PREDICTED: uncharacterized protein LOC105644073 [Jatropha curcas] |
| 12 |
Hb_001519_240 |
0.1869210788 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 13 |
Hb_029727_010 |
0.1885153086 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 14 |
Hb_007676_060 |
0.1887980265 |
- |
- |
PREDICTED: cellulose synthase-like protein E2 [Pyrus x bretschneideri] |
| 15 |
Hb_001288_050 |
0.1891427665 |
- |
- |
- |
| 16 |
Hb_000137_060 |
0.1898177631 |
- |
- |
hypothetical protein JCGZ_15115 [Jatropha curcas] |
| 17 |
Hb_169514_010 |
0.1905645157 |
- |
- |
PREDICTED: uncharacterized protein LOC103331076 [Prunus mume] |
| 18 |
Hb_001005_040 |
0.1917045927 |
- |
- |
- |
| 19 |
Hb_030885_010 |
0.1923207139 |
- |
- |
- |
| 20 |
Hb_006651_010 |
0.192416917 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4-like [Citrus sinensis] |