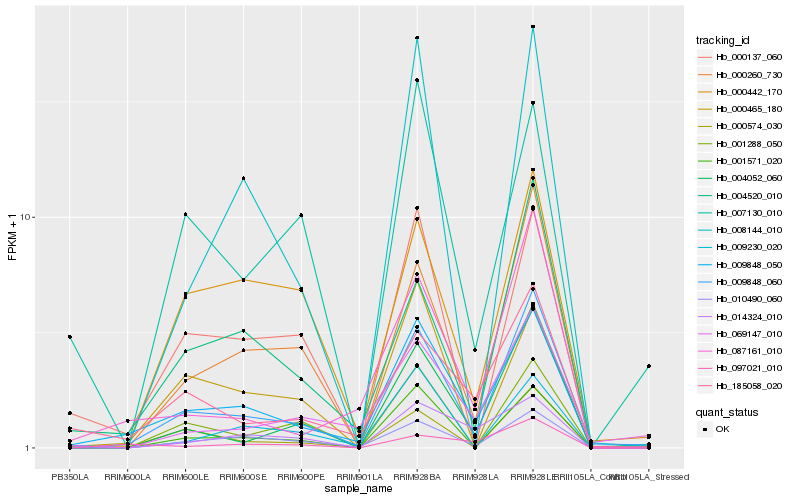
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009848_050 |
0.0 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK13 [Elaeis guineensis] |
| 2 |
Hb_009848_060 |
0.1506233811 |
- |
- |
unknown [Glycine max] |
| 3 |
Hb_010490_060 |
0.1657642002 |
- |
- |
PREDICTED: uncharacterized protein LOC105767021 [Gossypium raimondii] |
| 4 |
Hb_185058_020 |
0.1691951827 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 5 |
Hb_014324_010 |
0.1965704212 |
- |
- |
hypothetical protein JCGZ_05575 [Jatropha curcas] |
| 6 |
Hb_000137_060 |
0.1975457971 |
- |
- |
hypothetical protein JCGZ_15115 [Jatropha curcas] |
| 7 |
Hb_001571_020 |
0.1997232341 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850-like [Citrus sinensis] |
| 8 |
Hb_087161_010 |
0.2092480517 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_000465_180 |
0.2097262689 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 10 |
Hb_004520_010 |
0.2115629933 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 11 |
Hb_008144_010 |
0.2129111205 |
- |
- |
hypothetical protein VITISV_015004 [Vitis vinifera] |
| 12 |
Hb_000574_030 |
0.2134461697 |
desease resistance |
Gene Name: Glyco_hydro_38 |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 13 |
Hb_000260_730 |
0.2165672485 |
- |
- |
- |
| 14 |
Hb_069147_010 |
0.2177860615 |
desease resistance |
Gene Name: zf-BED |
PREDICTED: probable disease resistance protein At4g27220 isoform X1 [Populus euphratica] |
| 15 |
Hb_009230_020 |
0.2186324023 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000442_170 |
0.2199369939 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 17 |
Hb_097021_010 |
0.2238126698 |
- |
- |
hypothetical protein POPTR_0075s00200g, partial [Populus trichocarpa] |
| 18 |
Hb_004052_060 |
0.2246692247 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 19 |
Hb_007130_010 |
0.2252537846 |
- |
- |
- |
| 20 |
Hb_001288_050 |
0.2272489968 |
- |
- |
- |