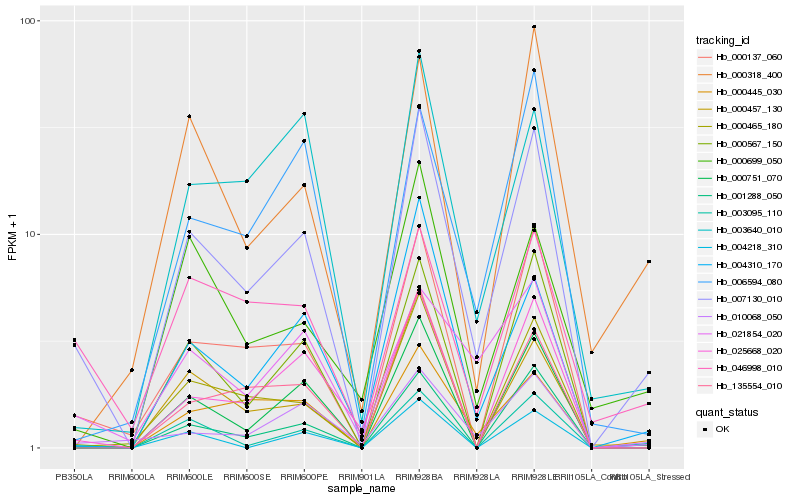
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007130_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_000137_060 |
0.1538438866 |
- |
- |
hypothetical protein JCGZ_15115 [Jatropha curcas] |
| 3 |
Hb_000567_150 |
0.1667928259 |
- |
- |
cysteine protease, putative [Ricinus communis] |
| 4 |
Hb_004310_170 |
0.1714028149 |
- |
- |
PREDICTED: protein mago nashi homolog [Jatropha curcas] |
| 5 |
Hb_000751_070 |
0.1747462682 |
- |
- |
- |
| 6 |
Hb_000465_180 |
0.1754177849 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001288_050 |
0.1787429439 |
- |
- |
- |
| 8 |
Hb_000445_030 |
0.1788166024 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase 5 [Jatropha curcas] |
| 9 |
Hb_010068_050 |
0.1799762034 |
- |
- |
PREDICTED: uncharacterized protein LOC105632963 [Jatropha curcas] |
| 10 |
Hb_004218_310 |
0.1859826649 |
- |
- |
PREDICTED: NADPH:quinone oxidoreductase-like [Camelina sativa] |
| 11 |
Hb_003095_110 |
0.1863814434 |
- |
- |
GTP-binding protein alpha subunit, gna, putative [Ricinus communis] |
| 12 |
Hb_025668_020 |
0.1866618982 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000457_130 |
0.1904581115 |
- |
- |
hypothetical protein AALP_AA3G009100 [Arabis alpina] |
| 14 |
Hb_000699_050 |
0.19582256 |
- |
- |
- |
| 15 |
Hb_135554_010 |
0.1963352578 |
- |
- |
Insulin-degrading enzyme, putative [Ricinus communis] |
| 16 |
Hb_003640_010 |
0.1980398714 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |
| 17 |
Hb_046998_010 |
0.1991295216 |
desease resistance |
Gene Name: LRR_4 |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 18 |
Hb_021854_020 |
0.199442052 |
- |
- |
hypothetical protein POPTR_1763s00200g, partial [Populus trichocarpa] |
| 19 |
Hb_006594_080 |
0.1995001677 |
- |
- |
beta-glucosidase, putative [Ricinus communis] |
| 20 |
Hb_000318_400 |
0.200506361 |
- |
- |
conserved hypothetical protein [Ricinus communis] |