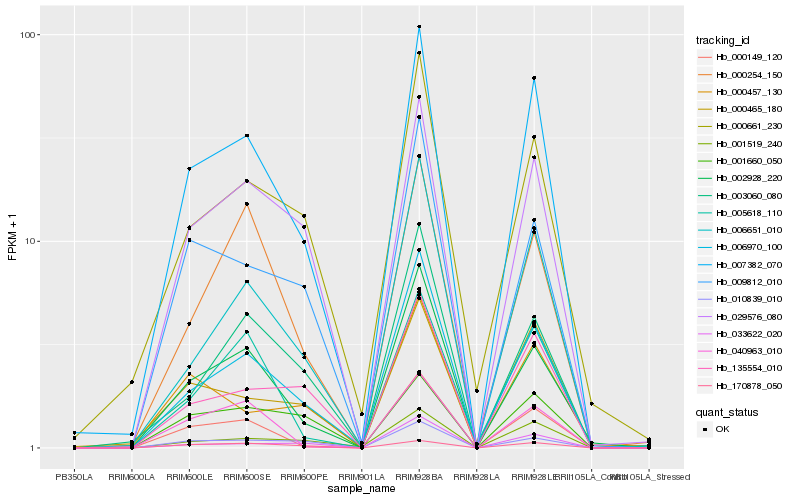
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007382_070 |
0.0 |
- |
- |
PREDICTED: MAPK-interacting and spindle-stabilizing protein-like [Jatropha curcas] |
| 2 |
Hb_006970_100 |
0.0937856516 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 3 |
Hb_000465_180 |
0.1113440614 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 4 |
Hb_029576_080 |
0.1137211666 |
- |
- |
Aspartic proteinase precursor, putative [Ricinus communis] |
| 5 |
Hb_002928_220 |
0.1189193723 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 6 |
Hb_000149_120 |
0.1201777318 |
- |
- |
PREDICTED: uncharacterized protein LOC105642119 isoform X2 [Jatropha curcas] |
| 7 |
Hb_006651_010 |
0.1223256124 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4-like [Citrus sinensis] |
| 8 |
Hb_001519_240 |
0.1242842026 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 9 |
Hb_135554_010 |
0.1257673469 |
- |
- |
Insulin-degrading enzyme, putative [Ricinus communis] |
| 10 |
Hb_000661_230 |
0.1380070522 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 22-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_009812_010 |
0.1403813363 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 12 |
Hb_005618_110 |
0.1472791153 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 13 |
Hb_010839_010 |
0.149254419 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 14 |
Hb_033622_020 |
0.1528628588 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 15 |
Hb_000457_130 |
0.1561381629 |
- |
- |
hypothetical protein AALP_AA3G009100 [Arabis alpina] |
| 16 |
Hb_000254_150 |
0.1564627804 |
- |
- |
PREDICTED: homeobox-leucine zipper protein GLABRA 2 [Jatropha curcas] |
| 17 |
Hb_003060_080 |
0.1568570406 |
- |
- |
PREDICTED: acyl-CoA--sterol O-acyltransferase 1 [Jatropha curcas] |
| 18 |
Hb_001660_050 |
0.1588310221 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Eucalyptus grandis] |
| 19 |
Hb_040963_010 |
0.1616040921 |
- |
- |
PREDICTED: polyneuridine-aldehyde esterase-like [Populus euphratica] |
| 20 |
Hb_170878_050 |
0.164602853 |
- |
- |
retrotransposon protein, putative, unclassified [Oryza sativa Japonica Group] |