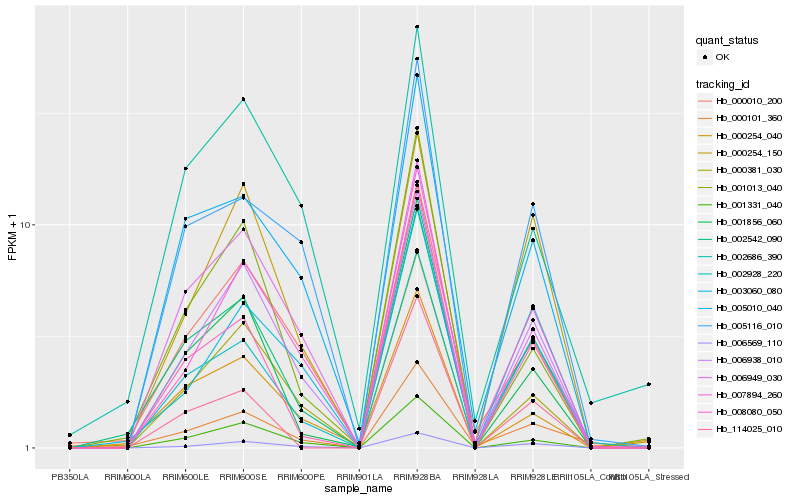
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002542_090 |
0.0 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 2 |
Hb_006938_010 |
0.1068881571 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 3 |
Hb_005010_040 |
0.1104898465 |
- |
- |
PREDICTED: uncharacterized protein LOC105630824 [Jatropha curcas] |
| 4 |
Hb_002928_220 |
0.1154867008 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 5 |
Hb_001331_040 |
0.1158740139 |
- |
- |
PREDICTED: cationic peroxidase 1-like [Jatropha curcas] |
| 6 |
Hb_000010_200 |
0.1230419854 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_001856_060 |
0.1231132286 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 8 |
Hb_000381_030 |
0.1248714294 |
- |
- |
PREDICTED: aquaporin SIP1-1-like [Jatropha curcas] |
| 9 |
Hb_006569_110 |
0.1291944709 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_006949_030 |
0.1315330441 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 11 |
Hb_000254_040 |
0.1330187924 |
- |
- |
PREDICTED: uncharacterized protein LOC105634992 [Jatropha curcas] |
| 12 |
Hb_007894_260 |
0.1356671889 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 13 |
Hb_000101_360 |
0.1367574022 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Jatropha curcas] |
| 14 |
Hb_001013_040 |
0.1377948673 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 51 [Jatropha curcas] |
| 15 |
Hb_008080_050 |
0.1389466006 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_005116_010 |
0.1449040244 |
transcription factor |
TF Family: bHLH |
PREDICTED: basic helix-loop-helix protein A [Jatropha curcas] |
| 17 |
Hb_000254_150 |
0.1555246565 |
- |
- |
PREDICTED: homeobox-leucine zipper protein GLABRA 2 [Jatropha curcas] |
| 18 |
Hb_114025_010 |
0.156488354 |
- |
- |
hypothetical protein POPTR_0008s13520g [Populus trichocarpa] |
| 19 |
Hb_003060_080 |
0.1570043626 |
- |
- |
PREDICTED: acyl-CoA--sterol O-acyltransferase 1 [Jatropha curcas] |
| 20 |
Hb_002686_390 |
0.1576251474 |
- |
- |
PREDICTED: uncharacterized protein LOC105647789 [Jatropha curcas] |