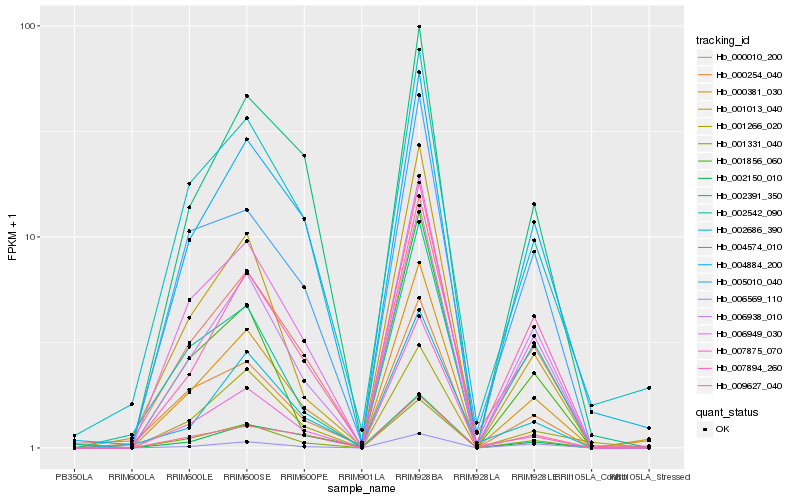
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000381_030 |
0.0 |
- |
- |
PREDICTED: aquaporin SIP1-1-like [Jatropha curcas] |
| 2 |
Hb_001331_040 |
0.0225173043 |
- |
- |
PREDICTED: cationic peroxidase 1-like [Jatropha curcas] |
| 3 |
Hb_006949_030 |
0.0693619568 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 4 |
Hb_006938_010 |
0.0749593852 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 5 |
Hb_000010_200 |
0.0934376502 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 6 |
Hb_001013_040 |
0.1020860268 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 51 [Jatropha curcas] |
| 7 |
Hb_007894_260 |
0.1021657486 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 8 |
Hb_007875_070 |
0.1131185081 |
- |
- |
hypothetical protein JCGZ_05017 [Jatropha curcas] |
| 9 |
Hb_004884_200 |
0.1141229356 |
- |
- |
PREDICTED: ferritin-3, chloroplastic [Jatropha curcas] |
| 10 |
Hb_005010_040 |
0.1167368028 |
- |
- |
PREDICTED: uncharacterized protein LOC105630824 [Jatropha curcas] |
| 11 |
Hb_002391_350 |
0.1184726368 |
- |
- |
PREDICTED: uncharacterized protein LOC105647080 [Jatropha curcas] |
| 12 |
Hb_006569_110 |
0.1197306705 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001856_060 |
0.1200532223 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 14 |
Hb_009627_040 |
0.1201822225 |
- |
- |
Lipase class 3-related protein, putative [Theobroma cacao] |
| 15 |
Hb_000254_040 |
0.1224067273 |
- |
- |
PREDICTED: uncharacterized protein LOC105634992 [Jatropha curcas] |
| 16 |
Hb_002150_010 |
0.1227332771 |
- |
- |
hypothetical protein POPTR_0001s10240g [Populus trichocarpa] |
| 17 |
Hb_002542_090 |
0.1248714294 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 18 |
Hb_002686_390 |
0.1264248685 |
- |
- |
PREDICTED: uncharacterized protein LOC105647789 [Jatropha curcas] |
| 19 |
Hb_004574_010 |
0.1287831413 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 20 |
Hb_001266_020 |
0.1297392377 |
- |
- |
PREDICTED: uncharacterized protein LOC105650395 [Jatropha curcas] |