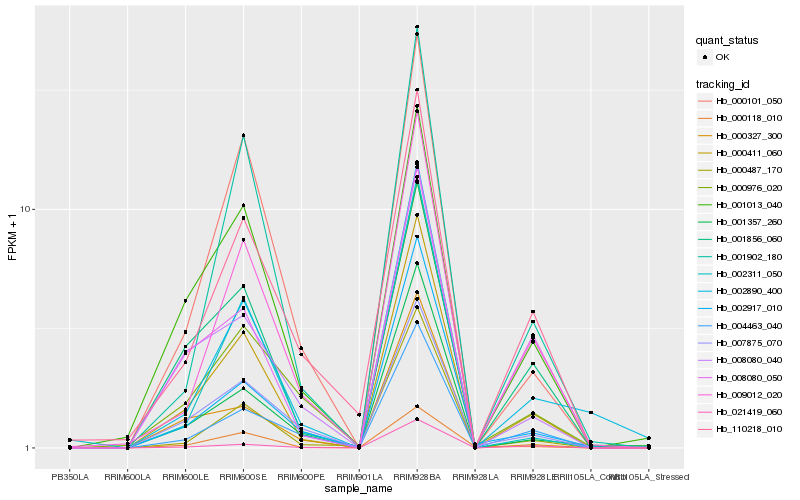
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000411_060 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 2 |
Hb_009012_020 |
0.103715681 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like [Populus euphratica] |
| 3 |
Hb_000487_170 |
0.1051312409 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 4 |
Hb_001902_180 |
0.1064143152 |
- |
- |
PREDICTED: uncharacterized protein LOC105129658 [Populus euphratica] |
| 5 |
Hb_000101_050 |
0.1065707056 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 6 |
Hb_008080_040 |
0.1128239978 |
- |
- |
hypothetical protein EUGRSUZ_D00834 [Eucalyptus grandis] |
| 7 |
Hb_001357_260 |
0.1181212543 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 8 |
Hb_001013_040 |
0.1181730053 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 51 [Jatropha curcas] |
| 9 |
Hb_004463_040 |
0.1183275768 |
- |
- |
- |
| 10 |
Hb_000976_020 |
0.118429432 |
- |
- |
leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 11 |
Hb_001856_060 |
0.1194464555 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 12 |
Hb_002311_050 |
0.1219902869 |
- |
- |
PREDICTED: uncharacterized protein LOC105122837 isoform X2 [Populus euphratica] |
| 13 |
Hb_000118_010 |
0.1221635491 |
desease resistance |
Gene Name: LRR_4 |
hypothetical protein POPTR_0021s00470g [Populus trichocarpa] |
| 14 |
Hb_002917_010 |
0.123098981 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 15 |
Hb_021419_060 |
0.1231584884 |
- |
- |
LRR receptor-like serine/threonine-protein kinase, putative [Theobroma cacao] |
| 16 |
Hb_002890_400 |
0.1272195011 |
- |
- |
2-nitropropane dioxygenase precursor, putative [Ricinus communis] |
| 17 |
Hb_110218_010 |
0.12737957 |
- |
- |
PREDICTED: uncharacterized protein LOC104428128 [Eucalyptus grandis] |
| 18 |
Hb_007875_070 |
0.128903684 |
- |
- |
hypothetical protein JCGZ_05017 [Jatropha curcas] |
| 19 |
Hb_008080_050 |
0.131982379 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_000327_300 |
0.1355252647 |
- |
- |
PREDICTED: ABC transporter G family member 6-like [Jatropha curcas] |