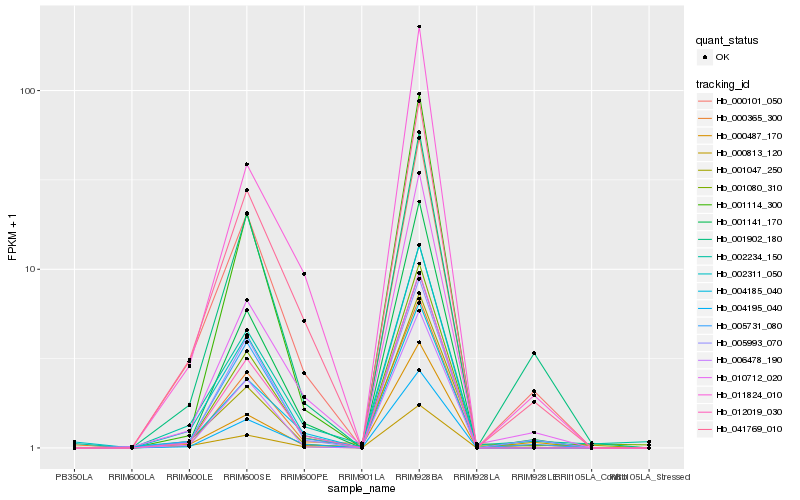
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002311_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105122837 isoform X2 [Populus euphratica] |
| 2 |
Hb_000365_300 |
0.0797031383 |
- |
- |
PREDICTED: cytochrome P450 714C2-like [Jatropha curcas] |
| 3 |
Hb_005731_080 |
0.0806467957 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 4 |
Hb_001141_170 |
0.0835053182 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At2g13820 [Jatropha curcas] |
| 5 |
Hb_012019_030 |
0.0855252477 |
- |
- |
hypothetical protein POPTR_0005s23360g [Populus trichocarpa] |
| 6 |
Hb_004195_040 |
0.0882633362 |
- |
- |
salicylic acid methyl transferase [Capsicum annuum] |
| 7 |
Hb_000813_120 |
0.0890240171 |
- |
- |
PREDICTED: serine carboxypeptidase-like 18 [Tarenaya hassleriana] |
| 8 |
Hb_002234_150 |
0.090261131 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 9 |
Hb_041769_010 |
0.0919963627 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 10 |
Hb_001080_310 |
0.0924823829 |
- |
- |
PREDICTED: ent-copalyl diphosphate synthase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000101_050 |
0.0991917693 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 12 |
Hb_006478_190 |
0.0996278451 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 13 |
Hb_004185_040 |
0.1010931363 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 14 |
Hb_005993_070 |
0.1032423598 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 15 |
Hb_010712_020 |
0.1042336955 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase-like [Jatropha curcas] |
| 16 |
Hb_001047_250 |
0.1055446001 |
- |
- |
PREDICTED: ent-copalyl diphosphate synthase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000487_170 |
0.106007673 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 18 |
Hb_001902_180 |
0.1073775695 |
- |
- |
PREDICTED: uncharacterized protein LOC105129658 [Populus euphratica] |
| 19 |
Hb_001114_300 |
0.1097474695 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 20 |
Hb_011824_010 |
0.1105296471 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |