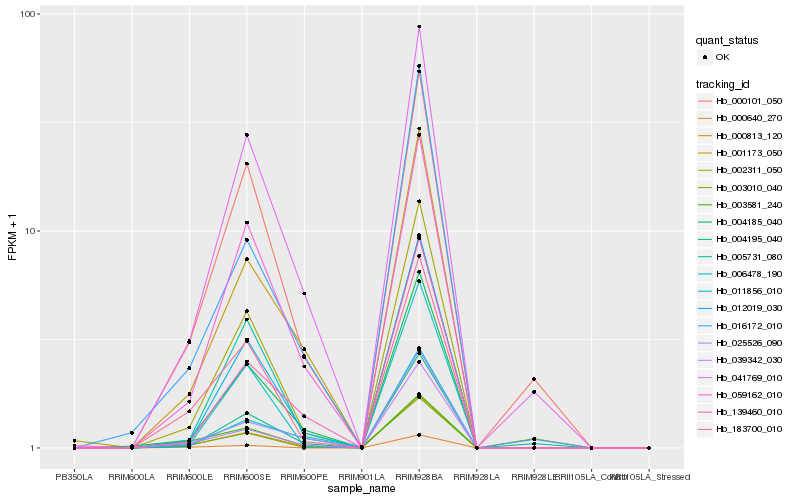
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000813_120 |
0.0 |
- |
- |
PREDICTED: serine carboxypeptidase-like 18 [Tarenaya hassleriana] |
| 2 |
Hb_183700_010 |
0.0706164165 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 3 |
Hb_039342_030 |
0.0761394088 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004185_040 |
0.0765824304 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 5 |
Hb_004195_040 |
0.0771479368 |
- |
- |
salicylic acid methyl transferase [Capsicum annuum] |
| 6 |
Hb_012019_030 |
0.0807909057 |
- |
- |
hypothetical protein POPTR_0005s23360g [Populus trichocarpa] |
| 7 |
Hb_001173_050 |
0.0870171084 |
- |
- |
DUF26 domain-containing protein 2 precursor, putative [Ricinus communis] |
| 8 |
Hb_003581_240 |
0.0878511192 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 9 |
Hb_002311_050 |
0.0890240171 |
- |
- |
PREDICTED: uncharacterized protein LOC105122837 isoform X2 [Populus euphratica] |
| 10 |
Hb_003010_040 |
0.0943523514 |
- |
- |
hypothetical protein AALP_AA7G248200 [Arabis alpina] |
| 11 |
Hb_041769_010 |
0.0944216696 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 12 |
Hb_016172_010 |
0.0996499788 |
- |
- |
hypothetical protein JCGZ_23602 [Jatropha curcas] |
| 13 |
Hb_059162_010 |
0.1005849231 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_000640_270 |
0.1015719273 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable pectinesterase 30 [Jatropha curcas] |
| 15 |
Hb_025526_090 |
0.1038836043 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 [Jatropha curcas] |
| 16 |
Hb_011856_010 |
0.1043211558 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 17 |
Hb_005731_080 |
0.1076926115 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 18 |
Hb_139460_010 |
0.1091408449 |
- |
- |
PREDICTED: beta-glucosidase 18-like [Jatropha curcas] |
| 19 |
Hb_006478_190 |
0.1097215046 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 20 |
Hb_000101_050 |
0.1105857175 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |