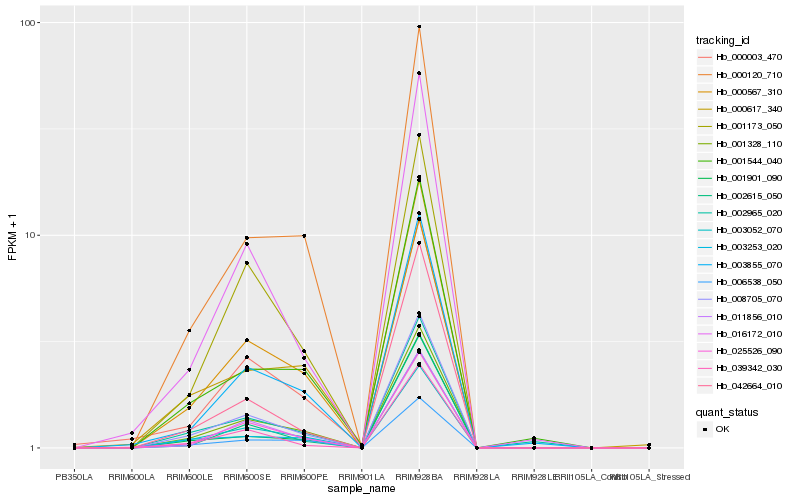
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008705_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638774 [Jatropha curcas] |
| 2 |
Hb_001328_110 |
0.0386220369 |
- |
- |
PREDICTED: LON peptidase N-terminal domain and RING finger protein 1-like [Jatropha curcas] |
| 3 |
Hb_039342_030 |
0.0686895865 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_025526_090 |
0.0693847675 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 [Jatropha curcas] |
| 5 |
Hb_003052_070 |
0.0705992321 |
- |
- |
PREDICTED: peroxidase 11 [Jatropha curcas] |
| 6 |
Hb_016172_010 |
0.0719655492 |
- |
- |
hypothetical protein JCGZ_23602 [Jatropha curcas] |
| 7 |
Hb_042664_010 |
0.0781597651 |
- |
- |
hypothetical protein POPTR_0001s07640g, partial [Populus trichocarpa] |
| 8 |
Hb_003253_020 |
0.078383938 |
transcription factor |
TF Family: B3 |
transcription factor, putative [Ricinus communis] |
| 9 |
Hb_002615_050 |
0.0874532729 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 10 |
Hb_001173_050 |
0.0882367616 |
- |
- |
DUF26 domain-containing protein 2 precursor, putative [Ricinus communis] |
| 11 |
Hb_000120_710 |
0.0883802138 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_011856_010 |
0.0889596479 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 13 |
Hb_003855_070 |
0.0893361106 |
- |
- |
hypothetical protein POPTR_0001s30980g [Populus trichocarpa] |
| 14 |
Hb_006538_050 |
0.0931632159 |
- |
- |
hypothetical protein RCOM_1308310 [Ricinus communis] |
| 15 |
Hb_000003_470 |
0.0943813243 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001901_090 |
0.0975440701 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 17 |
Hb_001544_040 |
0.0986902885 |
- |
- |
PREDICTED: uncharacterized protein At1g66480-like [Populus euphratica] |
| 18 |
Hb_002965_020 |
0.0992880631 |
- |
- |
hypothetical protein JCGZ_18947 [Jatropha curcas] |
| 19 |
Hb_000567_310 |
0.09968809 |
- |
- |
Miraculin precursor, putative [Ricinus communis] |
| 20 |
Hb_000617_340 |
0.1006471574 |
- |
- |
respiratory burst oxidase B [Manihot esculenta] |