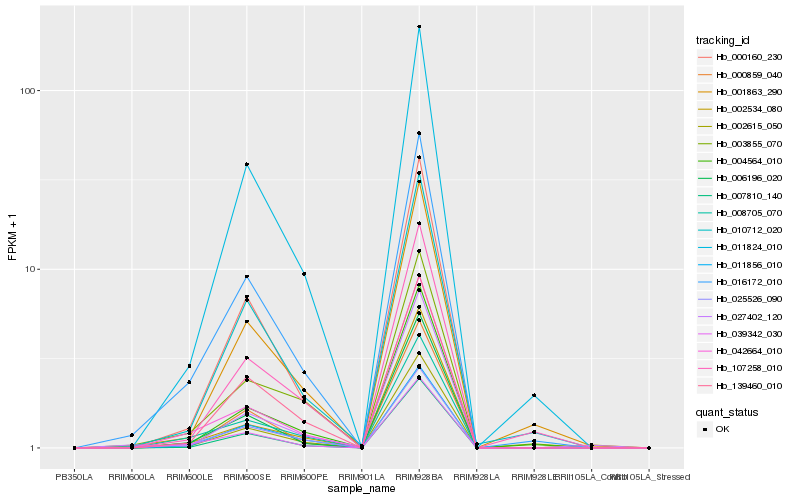
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002615_050 |
0.0 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 2 |
Hb_007810_140 |
0.0405801803 |
- |
- |
Ankyrin repeat-containing protein [Medicago truncatula] |
| 3 |
Hb_000160_230 |
0.0415001613 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 4 |
Hb_006196_020 |
0.053912388 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 100-like [Populus euphratica] |
| 5 |
Hb_011824_010 |
0.0598672433 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 6 |
Hb_000859_040 |
0.0613941248 |
- |
- |
Early nodulin 16 precursor, putative [Ricinus communis] |
| 7 |
Hb_139460_010 |
0.0629055013 |
- |
- |
PREDICTED: beta-glucosidase 18-like [Jatropha curcas] |
| 8 |
Hb_004564_010 |
0.0639415243 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 9 |
Hb_016172_010 |
0.0642457634 |
- |
- |
hypothetical protein JCGZ_23602 [Jatropha curcas] |
| 10 |
Hb_042664_010 |
0.0693876249 |
- |
- |
hypothetical protein POPTR_0001s07640g, partial [Populus trichocarpa] |
| 11 |
Hb_039342_030 |
0.0722870592 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_011856_010 |
0.0725172307 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 13 |
Hb_010712_020 |
0.0786105508 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase-like [Jatropha curcas] |
| 14 |
Hb_001863_290 |
0.0807782643 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 15 |
Hb_107258_010 |
0.083498397 |
- |
- |
hypothetical protein JCGZ_01141 [Jatropha curcas] |
| 16 |
Hb_008705_070 |
0.0874532729 |
- |
- |
PREDICTED: uncharacterized protein LOC105638774 [Jatropha curcas] |
| 17 |
Hb_002534_080 |
0.0899396502 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 18 |
Hb_025526_090 |
0.0918738365 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 [Jatropha curcas] |
| 19 |
Hb_003855_070 |
0.092885705 |
- |
- |
hypothetical protein POPTR_0001s30980g [Populus trichocarpa] |
| 20 |
Hb_027402_120 |
0.0944829982 |
- |
- |
zinc finger protein, putative [Ricinus communis] |