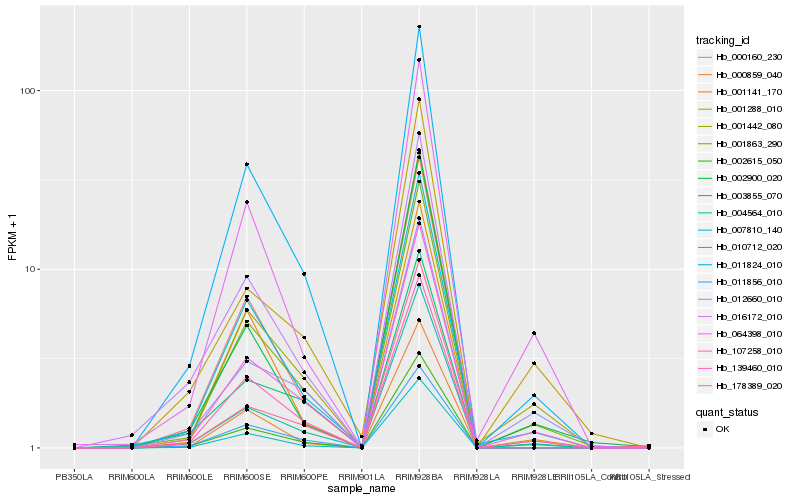
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001863_290 |
0.0 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 2 |
Hb_107258_010 |
0.0363521792 |
- |
- |
hypothetical protein JCGZ_01141 [Jatropha curcas] |
| 3 |
Hb_011824_010 |
0.0530671751 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 4 |
Hb_001442_080 |
0.0540814625 |
transcription factor |
TF Family: MYB-related |
transcription factor MYB251 [Fagus crenata] |
| 5 |
Hb_010712_020 |
0.0586801448 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase-like [Jatropha curcas] |
| 6 |
Hb_004564_010 |
0.06175558 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 7 |
Hb_064398_010 |
0.0710005146 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 8 |
Hb_012660_010 |
0.0801154883 |
- |
- |
hypothetical protein JCGZ_25971 [Jatropha curcas] |
| 9 |
Hb_002615_050 |
0.0807782643 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 10 |
Hb_007810_140 |
0.0853197264 |
- |
- |
Ankyrin repeat-containing protein [Medicago truncatula] |
| 11 |
Hb_000160_230 |
0.0873497194 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 12 |
Hb_016172_010 |
0.0883074206 |
- |
- |
hypothetical protein JCGZ_23602 [Jatropha curcas] |
| 13 |
Hb_139460_010 |
0.0884276511 |
- |
- |
PREDICTED: beta-glucosidase 18-like [Jatropha curcas] |
| 14 |
Hb_003855_070 |
0.088768346 |
- |
- |
hypothetical protein POPTR_0001s30980g [Populus trichocarpa] |
| 15 |
Hb_178389_020 |
0.0902564858 |
- |
- |
PREDICTED: cytochrome P450 86B1-like [Jatropha curcas] |
| 16 |
Hb_001288_010 |
0.091510298 |
- |
- |
- |
| 17 |
Hb_001141_170 |
0.0937854573 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At2g13820 [Jatropha curcas] |
| 18 |
Hb_000859_040 |
0.09449663 |
- |
- |
Early nodulin 16 precursor, putative [Ricinus communis] |
| 19 |
Hb_011856_010 |
0.0980082228 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_002900_020 |
0.0981535884 |
- |
- |
hypothetical protein POPTR_0008s06270g [Populus trichocarpa] |