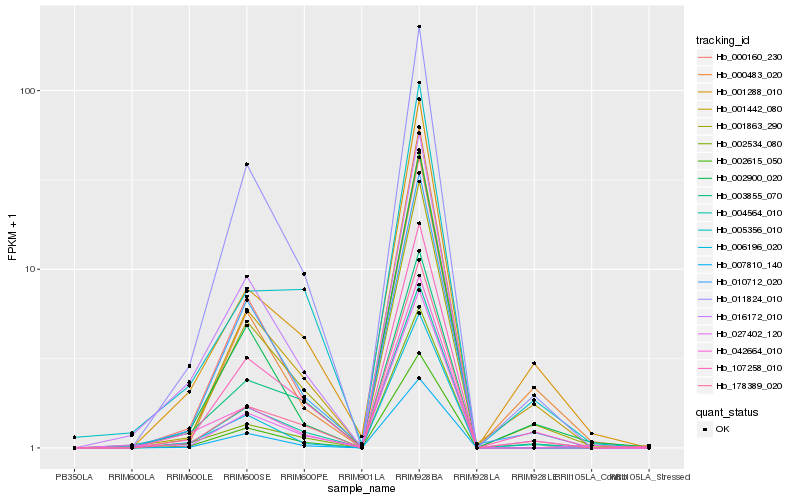
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004564_010 |
0.0 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 2 |
Hb_178389_020 |
0.0524733838 |
- |
- |
PREDICTED: cytochrome P450 86B1-like [Jatropha curcas] |
| 3 |
Hb_107258_010 |
0.0580504213 |
- |
- |
hypothetical protein JCGZ_01141 [Jatropha curcas] |
| 4 |
Hb_001442_080 |
0.0606455527 |
transcription factor |
TF Family: MYB-related |
transcription factor MYB251 [Fagus crenata] |
| 5 |
Hb_001863_290 |
0.06175558 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 6 |
Hb_002615_050 |
0.0639415243 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 7 |
Hb_001288_010 |
0.0725543856 |
- |
- |
- |
| 8 |
Hb_011824_010 |
0.0753819718 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_002900_020 |
0.0783006104 |
- |
- |
hypothetical protein POPTR_0008s06270g [Populus trichocarpa] |
| 10 |
Hb_010712_020 |
0.0821826472 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase-like [Jatropha curcas] |
| 11 |
Hb_007810_140 |
0.0823299617 |
- |
- |
Ankyrin repeat-containing protein [Medicago truncatula] |
| 12 |
Hb_000160_230 |
0.0823327724 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 13 |
Hb_006196_020 |
0.0833357464 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 100-like [Populus euphratica] |
| 14 |
Hb_042664_010 |
0.0842724832 |
- |
- |
hypothetical protein POPTR_0001s07640g, partial [Populus trichocarpa] |
| 15 |
Hb_016172_010 |
0.087704022 |
- |
- |
hypothetical protein JCGZ_23602 [Jatropha curcas] |
| 16 |
Hb_005356_010 |
0.0877151851 |
- |
- |
hypothetical protein POPTR_0003s16550g [Populus trichocarpa] |
| 17 |
Hb_003855_070 |
0.0897327538 |
- |
- |
hypothetical protein POPTR_0001s30980g [Populus trichocarpa] |
| 18 |
Hb_002534_080 |
0.0909948793 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 19 |
Hb_027402_120 |
0.0935293384 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 20 |
Hb_000483_020 |
0.0937786587 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |