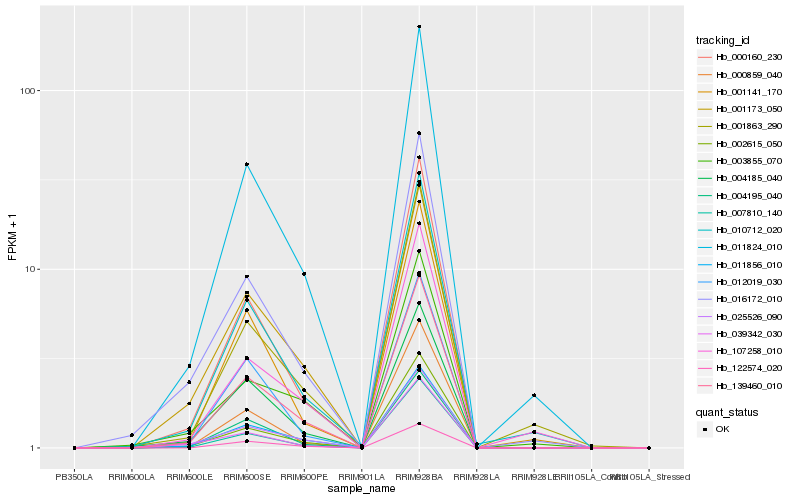
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_139460_010 |
0.0 |
- |
- |
PREDICTED: beta-glucosidase 18-like [Jatropha curcas] |
| 2 |
Hb_011856_010 |
0.0227717049 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 3 |
Hb_011824_010 |
0.0484231514 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 4 |
Hb_002615_050 |
0.0629055013 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 5 |
Hb_001173_050 |
0.0657086694 |
- |
- |
DUF26 domain-containing protein 2 precursor, putative [Ricinus communis] |
| 6 |
Hb_000160_230 |
0.0677216232 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 7 |
Hb_007810_140 |
0.0686117496 |
- |
- |
Ankyrin repeat-containing protein [Medicago truncatula] |
| 8 |
Hb_025526_090 |
0.070782958 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 [Jatropha curcas] |
| 9 |
Hb_004195_040 |
0.0729750971 |
- |
- |
salicylic acid methyl transferase [Capsicum annuum] |
| 10 |
Hb_122574_020 |
0.073872913 |
- |
- |
hypothetical protein JCGZ_24076 [Jatropha curcas] |
| 11 |
Hb_010712_020 |
0.0750541817 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase-like [Jatropha curcas] |
| 12 |
Hb_004185_040 |
0.0757217332 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 13 |
Hb_000859_040 |
0.0805598179 |
- |
- |
Early nodulin 16 precursor, putative [Ricinus communis] |
| 14 |
Hb_012019_030 |
0.0819247486 |
- |
- |
hypothetical protein POPTR_0005s23360g [Populus trichocarpa] |
| 15 |
Hb_016172_010 |
0.0835987153 |
- |
- |
hypothetical protein JCGZ_23602 [Jatropha curcas] |
| 16 |
Hb_001863_290 |
0.0884276511 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 17 |
Hb_107258_010 |
0.089742868 |
- |
- |
hypothetical protein JCGZ_01141 [Jatropha curcas] |
| 18 |
Hb_001141_170 |
0.0906708506 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At2g13820 [Jatropha curcas] |
| 19 |
Hb_003855_070 |
0.0966453978 |
- |
- |
hypothetical protein POPTR_0001s30980g [Populus trichocarpa] |
| 20 |
Hb_039342_030 |
0.0968848623 |
- |
- |
conserved hypothetical protein [Ricinus communis] |