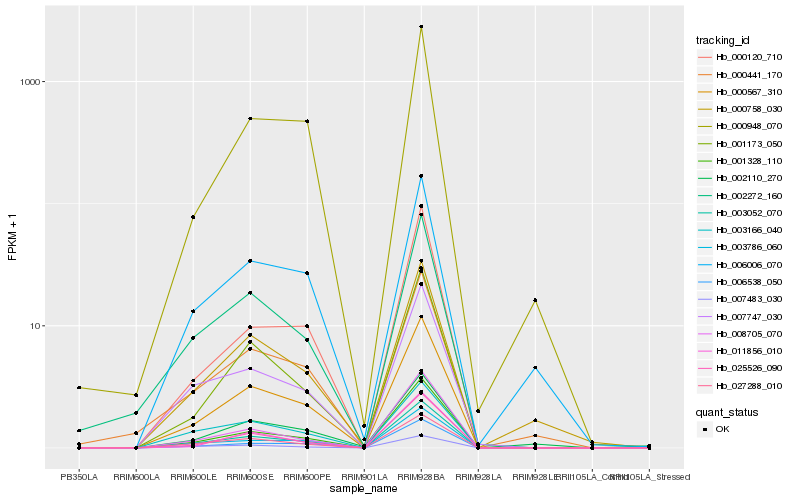
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000567_310 |
0.0 |
- |
- |
Miraculin precursor, putative [Ricinus communis] |
| 2 |
Hb_003052_070 |
0.0436255745 |
- |
- |
PREDICTED: peroxidase 11 [Jatropha curcas] |
| 3 |
Hb_006538_050 |
0.0676806272 |
- |
- |
hypothetical protein RCOM_1308310 [Ricinus communis] |
| 4 |
Hb_001328_110 |
0.0729142961 |
- |
- |
PREDICTED: LON peptidase N-terminal domain and RING finger protein 1-like [Jatropha curcas] |
| 5 |
Hb_027288_010 |
0.0732051646 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 6 |
Hb_001173_050 |
0.0745689368 |
- |
- |
DUF26 domain-containing protein 2 precursor, putative [Ricinus communis] |
| 7 |
Hb_003786_060 |
0.0793259523 |
- |
- |
PREDICTED: transmembrane protein 220 [Jatropha curcas] |
| 8 |
Hb_025526_090 |
0.0864847174 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 [Jatropha curcas] |
| 9 |
Hb_000948_070 |
0.0874153642 |
- |
- |
hypothetical protein POPTR_0016s01720g [Populus trichocarpa] |
| 10 |
Hb_007747_030 |
0.0923132808 |
- |
- |
PREDICTED: WD repeat-containing protein LWD2-like [Jatropha curcas] |
| 11 |
Hb_000441_170 |
0.0980580577 |
- |
- |
PREDICTED: uncharacterized protein LOC105636851 [Jatropha curcas] |
| 12 |
Hb_008705_070 |
0.09968809 |
- |
- |
PREDICTED: uncharacterized protein LOC105638774 [Jatropha curcas] |
| 13 |
Hb_000758_030 |
0.0999396115 |
transcription factor |
TF Family: ERF |
Ethylene-responsive transcription factor, putative [Ricinus communis] |
| 14 |
Hb_007483_030 |
0.101739817 |
- |
- |
- |
| 15 |
Hb_006006_070 |
0.1018473852 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 16 |
Hb_002272_160 |
0.1022323901 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_011856_010 |
0.1030946827 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 18 |
Hb_002110_270 |
0.1065235399 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH96 [Jatropha curcas] |
| 19 |
Hb_000120_710 |
0.1084336099 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_003166_040 |
0.1098109645 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.1 [Jatropha curcas] |